Introduction
Rice (Oryza sativa L.) is the main food for more than half of the world’s population. Globally, rice production is more than 470 million metric tons where India is contributing 264.77 million tons to the rice consuming population of the world.1 Major constraints like salinity limit the production of rice in highly cultivated regions like eastern part of India despite of ideal conditions like temperature and rainfall.
Rice plants to establish under salt stress conditions are crucial at germination and seedling stage development. Rice seed germination can determine its seedling growth and yield to some extent. In this direction, selection of high-yielding varieties for rapid and uniform germination under salt stress conditions can further contribute towards strengthening seedling vigor.2 The high-yielding rice varieties like MTU-7029, CR Boro dhan-2, MTU-1001, Pooja, Pratiksha, BPT-5204, Sarala, Jogesh, Lalat, Swarna sub-1, Nellore sannalu, Vamsi were selected for the present study. For instance, Lalat was developed and released in India based on its best performance at field level with yield ranging from 40-45 quintals/ha and resistant for many biotic stresses.3, 4 However, salinity-stress induced differential anatomical and molecular responses for these genotypes like Lalat at tribal dominated part of the country is far from being elucidated.
Salinity stress is negatively correlated to plant growth due to osmotic stress.5 The earlier and noticeable salt stress response is the reduction in leaf surface which shows protuberances followed by disruption of leaf blades as the stress intensifies.6 Consequently, ruptures the stomatal glands thereby reducing photosynthetic carbon-di-oxide (CO2) fixation leading to CO2 starvation. Furthermore, induces the production of osmoprotectants like proline protects chlorophyll molecules to certain severity of salt stress7 thereafter directing towards sodium (Na+) accumulation. In conjunction with such studies, reports on rice cultivars tend to possess active OsHKT1;5 and OsNHX1 alleles specific for remarkable tolerance to salt stress8,9,10 further expedited the research in identifying superior transcripts. As few novel transcripts associated with such salt-stress responsive alleles, a comprehensive study to relate physiological, biochemical, anatomical with transcriptional gene regulation analysis was performed using popular landraces.
The rice landraces used in the study are cultivated amongst tribal people at eastern part of India. Despite high-yielding and multiple resistances, these rice varieties3 possess high potential for crop improvement and they are still understudied for various other physiological, anatomical and molecular traits. Thus, it becomes scientist’s responsibility to explore genetically viable, high-yielding and stress tolerant rice varieties to the tribal society. To meet the demand of coastal tribal people through agriculture, this study has been initiated to explore the presently cultivated high-yielding rice landraces by understanding the association of physiological, biochemical and anatomical traits responsiveness with that of transcriptional gene regulation for salt tolerance.
Material and Methods
Plant Material
Rice seeds of 12 different varieties of Eastern part of India (MTU-7029, CR Boro dhan-2, MTU-1001, Pooja, Pratiksha, BPT-5204, Sarala, Jogesh, Lalat, Swarna sub-1, Nellore sannalu and Vamsi) were used in the study.
Physiological trait analysis
Germination percentage
The seeds used were healthy and uniform. Initially, germination percentage was calculated by sowing seeds (50 numbers) of each variety separately, in sterilized blotted petriplates. The seeds were sown and grown in the sterilized blotted petriplates and around 4 ml of sterile distilled water (control) and six different concentrations of salt (NaCl) solution (50 mM, 100 mM, 150 mM, 200 mM, 250 mM and 300 mM) was added. The replications (three) were maintained for each variety with different salt concentrations. The germination experiment was carried out in a chamber with constantly maintained temperature of 30ºC and were periodically checked and watered. The germination percentage was measured on alternative days till 11th day.
Fresh/Dry weight and Relative Water Content
After recording the germination rate, the seedlings of all genotypes were continued to grow till 14th day under 0 mM, 150 mM and 250 mM salt stress conditions. The fresh weight (FW) of the shoot part from each replication/variety was measured. They were transferred and immersed in sterile distilled water for 24 hrs. After the incubation time, turgor weight (TW) was recorded and dried the samples at 80°C for 3 days to measure dry weight (DW). Further, the values of fresh weight (FW), turgor weight (TW) and dry weight (DW) were used to calculate11 relative water content (RWC) as follows,
RWC = (Fresh Weight – Dry Weight)/(Turgid Weight – Dry Weight)*100
Biochemical parameters and analysis
Chlorophyll and Carotenoids estimation
The 14th day harvested seedlings (0.5 g) of all genotypes (control and salt-stressed) were homogenized in mortar and pestle separately by using 80 % acetone. The homogenized solution was centrifuged at 1500 – 2000 rpm for 8 – 10 min and the supernatant was collected. The total chlorophyll was measured at 663 nm and 645 nm and carotenoids at 520 nm.12 It was further calculated for content of chlorophyll a, chlorophyll b and total chlorophyll as follows,
Chl a = 12.7*(the value of 663 nm) – 2.69*(the value of 645 nm)
Chl b = 22.9*(the value of 645 nm) – 4.68*(the value of 663 nm)
Total Chl = 20.2*(the value of 645 nm) + 8.02*(the value of 663 nm)
The chlorophyll stability index (CSI) was analyzed according to Murty and Majumder13 and is expressed as a percentage
Leaf proline estimation
The control and NaCl treated 14th day old seedling samples of each variety were used to study the proline content.The samples were weighed uniformly to 0.5 g and homogenized in 1 ml of 3 % aqueous sulfo-salicylic acid. It was centrifuged at 8000 rpm for 12 min to remove the debris. A ratio of 1;1 (acid Ninhydrin and glacial acetic acid) was mixed with the supernatent and the mixture was heated to 100ºC in water bath for 1 hr and then the tubes were immediately transferred to ice bath. An equal aliquot of toluene was added to the mixture and vortexed for 15 to 20 sec. Further, the chromophore phase was used to measure the absorbance at 520 nm. The readings obtained were analyzed with a basic standard curve of L-Proline.14
Anatomical examinations
Scanning Electron Microscope
The two extreme salt responsive rice varieties i.e., Lalat (salt-tolerant) and MTU7029 (salt-sensitive) were selected. The seedlings (14th day old control and salt-treated samples/specimen) were measured 2 cm away from tip and 0.2 cm of the leaf part was cut laterally and processed separately. The leaf histology of extreme salt tolerant and sensitive rice leaf samples was observed using the Scanning electron microscope (JEOL 6510 LV Model, Japan). The leaf samples were processed to sterilize the surface and mount it in EMS Quantomix WET-SEM (QX-102) capsules on the stubs. The specimen was immersed in the QX imaging buffer for 5 to 10 minutes initially. The sample was not required to dry at critical point as it was mounted on to QX-102 capsules directly. The capsule was studded onto a metal stub with double sided carbon tape to fix it on the stage (Note: The QX-102 capsules are for imaging various wet biological samples and can be visualized directly without any coating or embedding) and the specimen was viewed under Scanning Electron Microscope (JEOL 6510 LV Model, Japan).
Molecular analysis
Polymerase chain reaction (PCR) based allele mining
The varieties Lalat (salt-tolerant) and MTU7029 (salt-sensitive) were selected for allele mining. The seedlings grown for 14 days were used to extract DNA by cetyl trimethyl ammonium bromide (CTAB) method as described.15 The extracted DNA was quantified using nano-drop (ND-1000 spectrophotometer) and its quality was assessed on ethidium bromide (EtBr) stained 0.8% (w/v) agarose gel electrophoresis. The salt-specific gene primers OsHKT1;5 and OsNHX1 were used to run polymerase chain reaction. The reaction mixtures contained OsHKT1;5 and OsNHX1 forward and reverse primers (5 pmoles) separately with 1x PCR buffer (Bangalore Genei, India), dNTPs (0.05 mM – Fermentas) and 1U of Taq DNA polymerase (Bangalore Genei, India) with 50 ng of DNA template to overall reaction volume of 10 μl. In the reaction, the template DNA was denatured at 94°C for 5 mins initially, annealed at 55°C for 30 sec and 72°C for 1min of primer extension followed by 35 cycles of PCR amplification with a final extension of 72°C for 7min. The amplified products were run electrophoretically, excised the embedded product and analysed to sequence. The sequences were analyzed in ‘PLACE’ and ‘PLANT CARE’ professional bioinformatics databases to identify cis-acting regulatory elements.16
Statistical Analysis
All recorded data were analyzed for analysis of variance (ANOVA) using the SPSS statistical software package version 20.0. The differences between treatments and genotypes were analyzed using Tukey’s Honest Significant Difference (HSD) test. Pearson’s correlation coefficients were used to draw inferences on relationships among various traits.
Results
Physiological response of rice varieties
A. Rate of Germination under salt stress conditions
The germination percentage was measured in all the genotypes (MTU-7029, CR Boro dhan-2, MTU-1001, Pooja, Pratiksha, BPT-5204, Sarala, Jogesh, Lalat, Swarna sub-1, Nellore sannalu and Vamsi), under different concentrations of salt (0, 50 mM, 100 mM, 150 mM, 200 mM 250 mM, 300 mM), to select two optimum levels of stress conditions. The rate of germination was measured on every alternative day and final germination percentage was calculated on 11th day. In all the genotypes, the germination rate decreased with increase in salinity stress concentrations. The pattern of germination rate was consistent under 50 mM salt stress among Lalat (96.33 %), Jogesh (94.66 %) and BPT 5204 (94.00 %). At 100 mM NaCl concentrations, the reduction over control (ROC) was more than 30 % among Pooja with 35 % and 38.66 % in MTU7029 and remaining all genotypes showed around 20 % ROC. The genotypes under 150 mM NaCl conditions exhibited 5 to 10 % difference reduction over 100 mM salt stress. The differences were highly significant at 250 mM salt stress conditions under which all the genotypes showed ≥50 % ROC except Lalat. A consistently similar degree of significance was noticed in Lalat with 33.66 % under 250 mM NaCl conditions. The genotype remained stable under NaCl concentration 300 mM with around ~3 % reduction over 250 mM stress conditions, whereas, all other genotypes with 100 % ROC. However, none of the genotypes survived at 300 mM NaCl stress conditions on 11th day except Lalat which showed 63.66 % germination rate. Hence, based on germination rate under seven different salt concentrations, three optimum NaCl treatments (0 mM, 150 mM and 250 mM) were selected. The physiological and biochemical level differentiations among these genotypes were further studied under 0 mM, 150 mM and 250 mM salt stress conditions.
B. Shoot fresh weight (FW) and dry weight (DW) to relate with relative water content (RWC)
Salinity had significant effect on shoot fresh weight (SFW) and dry weight (DW) of all the genotypes (P≤0.05) indicating increased salt concentrations decreased the fresh and dry weight. The shoot fresh weight among these genotypes against different salinity levels was significantly different. Lalat and Vamsi showed the highest SFW under control conditions and reduction over control (ROC) among these genotypes remained similar under high salt concentrations. Interestingly, a positive correlation was significant among the traits SFW, DW and RWC with respect to each genotype against the salt treatments (Table 1).
Table 1: Pearson’s correlation co-efficient amongst the physiological characteristics in genotypes sown and grown for 14 days under control, 150 and 250 mM NaCl at seedling stage.
|
FW |
DW |
RWC |
||
| FW |
1 |
|||
| DW |
0.639 |
1 |
||
| RWC |
0.470 |
0.223 |
1 |
Severe osmotic stress with marked dried shoot tips was observed in 14 days old seedlings. Relative water content (RWC) decreased significantly in all genotypes with increased salt stress conditions. Reduction over control (ROC) was noticed in all genotypes but the least reduction was significant in BPT 5204 with 4.26 % and Swarna sub-1 with 2.94 % difference from 150 mM to 250 mM salt concentrations. The results showed significant consistency in reduction among Lalat, Jogesh, Vamsi, Sarala and MTU1001 under both 150 and 250 mM salt stress conditions. A very high ROC in RWC was significant in Pooja (31.15 %) followed by MTU7029 (28.02 %) under 250 mM salt stress conditions.
Biochemical estimations
Chlorophyll and carotenoids content variations under salt stress conditions
A noticeable significant difference in chlorophyll content among all the genotypes over salinity treatment, within treatment as well as among genotypes was observed at P≤0.05 (Table 2). Total chlorophyll content decreased with increased salt concentrations. A minimum reduction over control (ROC) was significant in Lalat with 1.3 folds and maximum ROC in Vamsi under 150 mM NaCl stress conditions. The performance was visibly poor and evident in all the genotypes with ROC more than ~5.0 folds under 250 mM NaCl stress conditions, except for Lalat. The traces of chlorophyll were slightly significant among BPT 5204, Sarala and CR Boro dhan-2 with ROC of 4.3, 6.6 and 8.3 folds respectively under 250 mM stress conditions. The level of carotenoids in all the genotypes against salt stress conditions was highly significant at P≤0.01. The high carotenoid content was prominent under 150 mM salt stress conditions in MTU1001 followed by Swarna sub-1, Pratiksha, Sarala, CR Boro dhan-2 and Lalat. On the contrary, the reduction in carotenoid level was sharp among most of the genotypes at 250 mM salt stress conditions. The genotypes Sarala, CR Boro dhan-2, Pratiksha, Pooja, BPT 5204, Swarna sub-1, MTU1001, Jogesh and Nellore sannalu showed significant reduction of carotenoids under 250 mM salt stress conditions. However, Lalat remained consistent throughout even under higher salt stress conditions and a total contrasting performance was recorded in MTU7029 as well as Vamsi under 250 mM NaCl treatments for carotenoid content.
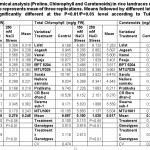 |
Table 2: Biochemical analysis (Proline, Chlorophyll and Carotenoids) in rice landraces of Eastern part of India. Each value represents mean of three replications. Means followed by different letters in the same column are significantly different at the P<0.01/P<0.05 level according to Tukey’s HSD test. Click here to View table |
A. Proline accumulations and its variations
Increasing salt concentrations triggered the level of proline in leaves of all the genotypes used. The proline content increased with increasing salt concentrations in all the genotypes except for Pooja, Sarala, Pratiksha and MTU1001. These genotypes exhibited a trend of increased proline at 150 mM NaCl over control and decreased trend at 250 mM salt stress conditions. The genotypes including Lalat, Jogesh, Vamsi, BPT 5204, MTU7029, Nellore sannalu and CR Boro dhan-2 showed a noticeable increase in proline content with increasing salt concentrations. The increase of proline in leaves of all genotypes was more than 100 % in 150 mM NaCl over control conditions. Despite of increased proline content under 150 mM salt stress conditions, Swarna sub-1 demonstrated consistency in its level of proline even at 250 mM salt treatment. A highly significant variation at P≤0.05 was observed among the treatments, genotypes and genotype over treatment for proline content at seedling stage (Table 2). Invariably, the performance of Lalat was highly significant and found to be the most salt-tolerant under higher concentrations of salinity for all the traits measured, whereas, the performance of MTU7029 was significantly low under salt stress conditions. Therefore, the control and 150 mM salt treatments were further short-listed to study the anatomical and molecular traits of two-contrasting genotypes more precisely.
Anatomical changes in rice genotypes under salt stress
The cell damage on the adaxial surface of rice leaves due to salinity treatments were studied using scanning electron microscope. The study showed very less opened glands on the leaves of 14 days old seedlings of Lalat and MTU7029 under control conditions (Figure No.: 1A, C). However, the glands opened and adaxial surface destructed the leaf blade of Lalat and MTU7029 under 14 days old 150 mM NaCl treated seedlings (Figure No.: 1B, D). The surface of Lalat leaf exhibited less bulging and opened stomatal glands in response to salt stress (Figure No.: 1B). In comparison, a significantly noticeable and large number of swollen as well as opened glands were prominent in adaxial leaf surface of MTU7029 (Figure No.: 1D). Despite of stomatal gland bulging appeared on leaves, no ruptures were prominent on the surface of the adaxial leaf blade under 150 mM NaCl stress conditions. Hence, the anatomical studies on rice adaxial leaf surfaces demonstrated significant differences between two contrasting landraces in response to salt stress.
 |
Figure 1: Scanning electron microscopy of adaxial leaf surface of rice seedlings. Click here to View figure |
Scanning electron micrographs 1: (A) Adaxial side of Lalat (salt-tolerant) leaf under control conditions (B) Adaxial side of Lalat (salt-tolerant) leaf under 150 mM NaCl conditions (C) Adaxial side of MTU 7029 (salt-sensitive) leaf under Control conditions (D) Adaxial side of MTU 7029 (salt-sensitive) leaf under 150 mM NaCl conditions.
Molecular level responses of rice landraces
Allelic variations and stress-responsive superior cis-acting regulatory elements
The primer OsHKT1;5 and OsNHX1 was used spanning both exon and intron set with amplicon sizes 1.6 kb and 1.5 kb respectively. Among these set of primers, significant variations in the molecular weight of the amplified products were detected among both the genotypes (Lalat and MTU7029). A noticeable variation was apparent in both the genotypes with primers OsHKT1;5 and OsNHX1 (Figure No.: 2). Hence, allelic variations distinguished among both the genotypes with mentioned primers were subjected to sequencing and subsequent in silico bioinformatic analysis. The analysis was carried out for potential cis-acting regulatory elements (CARE) to identify nucleotide sequence motifs. Possible cis-acting regulatory elements were identified in the 5` regulatory sequences of the genotypes Lalat and MTU7029 (Supplementary Figure No.: 1 to 4). Despite of few common cis-acting regulatory elements (CARE) among both the genotypes, several superior salt-stress responsive motifs were identified mostly in salt-tolerant Lalat.
 |
Figure 2: Polymerase chain reaction for OsNHX1 and OsHKT1;5 in Lalat and MTU7029. Click here to View figure |
The PCR-based allele mining shows allelic variations between the two-contrasting rice genotypes Lalat (salt-tolerant) and MTU7029 (salt-sensitive) using the salt-specific homologous primers OsNHX1 and OsHKT1;5 with two distinct variations.
When analysed with OsNHX1, various motifs were found which are represented here in Supplementary figures. The bioinformatics analysis was conducted for two-contrasting genotypes i.e., salt-tolerant Lalat and salt-sensitive MTU7029, to further explore the understanding at molecular level. The motifs CAAT, Skn-1 and TATA-box were common among both the genotypes with OsNHX1. Several light responsive cis-acting regulatory elements like ATCT-motif, CATT-motif, GAG-motif and Sp1 were present in Lalat at various 5` regulatory region with CAAT-box, which is a common CARE in promoter and enhancer region (Supplementary Figure No.: 1A to 1D). Notably, it is very important to observe in salt-tolerant Lalat that superior motifs Box-W1, MBS and W-box were associated with fungal elicitor responsive element, MYB binding site specific for abiotic stress response and salt-stress responsiveness, respectively. In addition to all the mentioned cis-acting regulatory elements, several hormonal responsive motifs such as TCA element, TGA element and CGTCA/TGACG-motifs specific for salicylic acid, auxin and meJA-responsiveness were identified in salt-tolerant genotype. The sequential information map of Lalat was a reference for salt-sensitive MTU7029 with OsNHX1 gene. The motifs HSE and LTR significantly present in MTU7029 (salt-sensitive) genotype which are specific for high and low-temperatures respectively (Supplementary Figure No.: 2A and 2B). There was no salt-stress responsive CAREs identified in MTU7029. Apart from possessing such cis-regulatory elements, both the genotypes included few unnamed elements like unnamed-4, -3, etc., which are associated with methylation protection and immune-precipitation.
Analysis of the 5` regulatory regions of the sequences using OsHKT1;5 in the salt-tolerant Lalat revealed superior motifs E2Fb and GC motif in addition to Box-W1, W box and MBS (Supplementary Figure No.: 3A to 3C). The transcription activator E2Fb stimulates cell proliferation and delays differentiation and GC-motif involved in stress responsiveness. On the contrary, the gene OsHKT1;5 in salt-sensitive MTU7029 showed no significant motifs except for CAAT, TATA-box, GARE, LTR and Skn-1 (Supplementary Figure No.: 4A and 4B). Overall, salt-tolerant Lalat performed best suggesting possible involvement of salt-stress responsive superior transcripts.
Discussion
The germination test can determine the percentage of seeds that are viable in a given sample. The degree of germination in relation with seed vigour estimates its potential field performance.17 The seed germination is dependent on seed structure as well as the environmental factors that affect the growth of the embryo. In the present study, the germination rate in most of the genotypes was inhibited at higher concentrations of salinity stress.18, 19 Salinity stress had significant effect on all the genotypes on their germination rate. The reduction over control was high in MTU-7029, CR Boro dhan-2, MTU-1001, Pooja, Pratiksha, BPT-5204, Sarala, Jogesh, Swarna sub-1, Nellore sannalu and Vamsi except Lalat under different concentrations of salt like 0, 50 mM, 100 mM, 150 mM, 200 mM 250 mM, 300 mM. The germination rate remained invariable in Lalat but none of other genotypes survived. Hence, the study was further focused with salt treatments 0, 150 and 250 mM to concretize the structure of physiological and biochemical traits.
In this direction, few physiological traits like fresh weight (FW), dry weight (DW) and relative water content (RWC) was measured in all the genotypes. A significant positive correlation was observed between FW, DW and RWC.20, 21, 22 and 23 Relative water content is a potential parameter to quantify the degree of osmotic stress in plants.24 The reduction over control of RWC was significant under 150 mM NaCl stress conditions but significantly high reduction was recorded with increased salt stress conditions. A similar trend of reduction over control in RWC was prominent in most of the genotypes, indicating higher level of reduction with increased degree of stress.
Proline content in leaves of all the genotypes increased with increasing salt concentrations. A similar observation was reported in other parts of tomato plants like stem and leaves under 160 mM NaCl25 treatments. The trend remained similar to proline content among introgression lines derived from wild types of rice.26 In the present study, the long term exposure to NaCl stress exhibited ~six folds increase in the most salt-tolerant Lalat whereas ~five folds in the most salt-sensitive MTU7029 (Table No.: 2). Proline, an osmoprotectant gets accumulated as an important defense mechanism in higher plants. The accumulation is at higher level under stress conditions like salinity, drought, cold, etc., and it also protects the photosynthetic pigment chlorophyll. The chlorophyll pigments are more sensitive to salinity stress in salt sensitive rice genotypes27 due to hyper-sensitivity of chlorophyll-b to salinity than that of chlorophyll-a.28 Though, some of the genotypes in the present study showed highest proline content, but their performance for other biochemical traits was poor. For instance, Vamsi indicated with highest proline content (~30 folds) under high concentrations of salinity (250 mM) but with lowest chlorophyll as well as carotenoid content. However, Lalat performed best among all the genotypes with consistency in all the traits like chlorophyll and carotenoids. It was only ~2 folds reduction under stress conditions over control. Under salt stress conditions, many rice genotypes were found to show increased proline content with increasing salinity levels.26, 29, 30 and 31
Salinity stress adversely influences the mechanism of stomatal conductance in plants by disrupting the glands/pores.32 and 33 In the current study, an attempt of understanding the anatomy of stomatal glands under salt stress conditions resulted in the significantly affected leaf blades on the adaxial surface of rice seedlings of salt-sensitive genotype. The stomatal glands on adaxial leaf surface were uniform throughout in both the genotypes under control conditions but the glands bulged and widened intensely in salt-sensitive MTU7029 under salt stress conditions. The effect was remarkably less in the leaf blades of salt-tolerant Lalat. The stomatal glands regulate the uptake of carbon-di-oxide for the process of photosynthesis against loss of water but under salt stress conditions, the glands increase in number and gets widened leading to dryness of leaves by inhibiting metabolic rate in plants.31
Diversified genetic information is one of the sources at molecular level to screen and shortlist the genotypes for various stress tolerance. Usage of trait specific markers for phenotyping would expedite the process of genotypic selection for allele mining. Several molecular and bioinformatic tools like PCR-allele mining would expedite the identification of superior alleles and nucleotide sequence motifs. Various studies have been reported on characterization of genes,34, 35 but very limited is known about the cis-acting regulatory elements and their regulation in plants involved in stress-responsive expressions in plants.36 The genomic sequence information of Arabidopsis/rice37, 38 and 39 and professional bioinformatic tools have unfastened the way for advanced studies on developmental, regulation and expression of novel genes. Significant variations in PCR-based allele mining and identification of cis-acting regulatory elements were observed in both salt-tolerant Lalat and salt-sensitive MTU7029 genotypes. All the sequences obtained from allele-mining were used for identification of cis-acting regulatory elements. These elements involved in gene expression and some in regulatory mechanisms that contribute to stress-regulation in rice genotypes.40 Many motifs are associated with complex protein domains and transcription factors coding for multiple regulatory mechanisms.41 Interestingly, salt-tolerant Lalat exhibited regulatory elements that are associated with salt stress-responsiveness. The regulatory elements like Skn-1, CAAT and TAAT-box in OsNHX1 were common in both the genotypes that are required for endosperm expression, in promoter and enhancer regions and around -30 of transcription start respectively and the result was similar to some of the studies reported in aquatic plants.42,43 Studies reported a salt-responsive BADH promoter from Atriplex centralasiatica and found elements like GC-motif and CAAT-box related to stress.44 and 45 The 5` untranslated region (5` UTR) of OsNHX1 contained superior motifs Box-W1, MBS and W-box associated with WRKY factors that mainly bind to one potential W-box that determine distinct transcriptional outputs.46 Stress response associated motifs like HSR and LTR were significant in salt-sensitive MTU7029 but not specific to salinity stress. The cis-regulatory elements like MYB binding site motifs are noticed in Lalat associated with abiotic stresses like drought and salinity and similar results were reported recently in Arabidopsis.47, 48
With the gene OsHKT1;5 in Lalat, more salt-stress specific motifs W-boxes, CAAT-box, E2Fb, G-box, GC-motif and MBS were found. The G-boxes are known to get regulated under various stresses such as UV irridation, salt stresses, hormonal imbalances, etc.49 The superior motif E2Fb is a transcription activator that is involved in cell cycle regulation or in DNA replication by stimulating cell proliferation and delays differentiation.50 Hence, PCR-based allele mining in our study demonstrated transcriptional regulation of the stress-responsive genes and superior alleles in Lalat, confirming the genotype as salt-tolerant at molecular level.
Conclusions
We characterized the rice landraces at physiological, biochemical, anatomical and molecular level at seedling stage. Our results revealed the dynamic invariable performance of genotypes under different and high concentrations of salt. The changes in expression patterns of all genotypes demonstrated in various traits facilitated in selecting extreme salt-tolerant and salt-sensitive landraces. The physiological and biochemical traits revealed Lalat as the best performer with high proline content that in turn protected the levels of chlorophyll even under high salinity stress conditions. The anatomical structure of adaxial leaf surface also indicated less bulging and few numbers of widely opened stomatal glands in Lalat on contrary to salt-sensitive MTU7029. All the traits correlated positively with allele mining variations in salt-specific genes OsNHX1 and OsHKT1;5 among Lalat and MTU7029 genotypes. The study also disclosed the involvement of probable and potential association of cis-acting regulatory elements in expression and regulation of salt-stress responsiveness in these two-contrasting genotypes. This strategy has a constructive foundation to study the organizational mechanisms for gene regulations and transcriptional association with stress-responsive superior alleles in various rice genotypes.
Acknowledgments
The authors sincerely acknowledge Prof. D. N. Rao, Vice-President, CUTM, Odisha for his encouragement and uninterrupted co-operation to conduct this major research part of the work in the manuscript at different departments of the university.
References
- Agricultural statistics at a Glance. Government of India, Ministry of Agriculture, Department of Agriculture and Cooperation, Directorate of Economics and Statistics. ISBN-13:978-0-19-945965-0 ISBN-10: 0-19-945965-7: (2014).
- Ueda A, Kathiresan A, Inada M, et al. Osmotic stress in barley regulates expression of a different set of genes than salt stress does. Journal of Experimental Botany; 406: 2213-2218: (2004).
CrossRef - Das S. R, Rice in Odisha, International Rice Research Institute (IRRI), Technical Bulletin; 16: (2012).
- DRR (Directorate of Rice Research), System of rice intensification: Enhancing input use efficiency in rice, DRR Technical Bulletin; 58: 27: (2011).
- Munns R. Genes and salt tolerance: bringing them together. New Phytologist; 167: 645-663: (2005).
CrossRef - Parida A. K, Das A. B. Salt tolerance and salinity effects on plants: A review. Ecotoxicology and Environmental Safety; 60(3): 324–349: (2005).
CrossRef - Wu J. T, Hsieh M. T, Kow L. C. Role of proline accumulation in response to toxic copper in Chlorella sp. (chlorophyceae) cells. Journal of Phycology; 34: 113-117: (1998).
CrossRef - Fukuda A, Nakamura A, Hara N, et al. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta; 233(1): 175-188: (2011).
CrossRef - Liu S, Zheng L, Xue Y, et al. Over-expression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. Journal of Plant Biology; 53(6): 444-452: (2010).
CrossRef - Ren Z. H, Gao J. P, Li L. G, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics; 37: 1141-1146: (2005).
CrossRef - Pushpalatha G, Subrahmanyam D, Sreenu K, et al. Effect of salt stress on seedling growth and antioxidant enzymes in two contrasting rice introgression lines. Indian Journal of Plant Physiology; 18(4): 360–366: (2013).
CrossRef - Palta J. P. Stress interactions at the cellular and membrane levels. Horticultural Science; 25: 1377-1381: (1990).
- Murty K. S, Majumder S. K. Modifications of technique for determination of chlorophyll stability index in relation to studies of drought resistance in rice. Current Science; 31: 470-471: (1962).
- Bates L. S, Waldren R. P, Teare I. D. Rapid determination of free proline for water stress studies. Plant Soil; 39: 205 -207: (1973).
CrossRef - Zheng K. L, Huang N, Bennett J, Khush G. S. PCR marker-assisted selection in rice breeding. IRRI Discussion Paper Series No.12: 1–24: IRRI, Manila, Philippines: (1995).
- Higo K, Ugawa Y, Iwamoto M. Plants cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research; 27(1): 297-300: (1999).
CrossRef - IRRI Rice Knowledge Bank. A note by M Gummert with inputs from J. F. Rickman. http://irri.org/news/119-wild-parent-spawns-super-salt-tolerant-rice (Accessed 15.04.2013): (2013).
- Khanam M, Al-Yeasa M, Sazzadur-Rahman Md, Al-Mahbub A, Gomosta A. R. Effects of different factors on the growth efficiency of rice seedlings. Bangladesh Journal of Botany; 36(2): 171–176: (2007).
- Ologundudu A. F, Adelusi A. A, Akinwale R. O. Effect of salt stress on germination and growth parameters of rice (Oryza sativa L.). Notulae Scientia Biologicae; 6(2): 237-243: (2014).
CrossRef - Tanveer U. H, Javaid A, John G, et al. Genetic mapping of QTLs, controlling shoot fresh and dry weight under salt stress in rice (Oryza sativa L.) cross between CO39 × Moroberekan. Pakistan Journal of Botany; 40(6): 2369-2381: (2008).
- Koyama M. L, Levesley A, Koebner R. M. D, et al. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiology; 125: 406-422: (2001).
CrossRef - Xiao J, Li J, Yuan L, et al. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from specific rice cross. Theoretical and Applied Genetics; 92: 230-244: (1996).
CrossRef - Flowers T. J, Yeo A. R. Breeding for salinity resistance in crop plants: where next? Australian Journal of Plant Physiology; 22: 875-884: (1995).
CrossRef - Niu G, Cabrera R. I. Growth and physiological responses of landscape plants to saline water irrigation: A review. Hortscience; 45(11): (2010).
- Amini F, Ali A. E. Soluble proteins, proline, carbohydrates and Na+/K+ changes in two tomato (Lycopersicon esculentum Mill.) cultivars under in vitro salt stress. American Journal of Biochemistry and Biotechnology; 1(4): 204-208: (2005).
CrossRef - Pushpalatha G, Giri A, Sarla N, et al. Comparative biochemical analysis of wild introgression lines in response to short-term exposure to salinity. Asian Journal of BioScience; 9(1): 1-8: (2014).
- Turan M. A, Awad Elkarim A. H, Taban N, Taban S. Effect of salt stress on growth, stomatal resistance, proline and chlorophyll concentrations on maize plant. African Journal of Agricultural Research; 4(9): 893–897: (2009).
- Doganlar Z. B, Demir K, Basak H, et al. Effects of salt stress on pigment and total soluble protein contents of three different tomato cultivars. African Journal of Agricultural Research; 5(15): 2056–2065: (2010).
- Zahra S, Amin B, Ali V. S. M, et al. The salicylic acid effect on the tomato (Lycopersicum esculentum Mill.) sugar, protein and proline contents under salinity stress (NaCl). Journal of Biophysics and Structural Biology; 2(3): 35–41: (2010).
- Chutipaijit S, Chaum S, Sompornpailin K. Differential accumulations of proline and flavonoids in Indica rice varieties against salinity. Pakistan Journal of Botany; 41(5): 2497–2506: (2009).
- Moradi F, Ismail A. M. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Annals of Botany; 99(6): 1161–1173: (2007).
CrossRef - Gomes D. M. S, Neves L. J. Scanning electron microscopy of the leaf epidermis of Merostachys Spreng (Poaceae: Bambusoideae). Acta Botanica Brasilica; 23(2): 516-525: (2009).
CrossRef - Alam M. A, Juraimi A. S, Rafii M. Y, et al. Effect of salinity on biomass yield and physiological and stem-root anatomical characteristics of purslane (portulaca oleracea l.) accessions. Hindawi publishing corporation, BioMed Research International; 15: (2015).
- Aoki N, Hirose T, Scofield G. N, et al. The sucrose transporter gene family in rice. Plant Cell Physiology; 44: 223-232: (2003).
CrossRef - Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Letters; 581: 2309-2317: (2007).
CrossRef - Meyer S, Lauterbach C, Niedermeier M, et al. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiology; 134: 684-693: (2004).
CrossRef - Zhao H, Wen Y, Yidan O, et al. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Research; 43: D1018–D1022: (2015).
CrossRef - Yu J, Hu S, Wang J, et al. A draft sequence of the rice genome (Orysa sativa L. ssp. Indica). Science; 296: 79-92: (2002).
CrossRef - Arabidopsis genome initiative – Arabidopsis information resources TAIR;https://www.arabidopsis.org/search/ERwin/Tair.htm: (2000).
- Qin D, Wu H, Peng H, et al. Heat-stress responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivaum L.) by using wheat genome array. BMC Genomics; 9: 432: (2008).
CrossRef - Yanagisawa S. The Dof family of plant transcription factors – A Review. Trends in Plant Science; 7(12): 555-560: (2002).
CrossRef - Kusnetsov V, Landsberger M, Meurer J, et al. The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the Plastids. Biological Chemistry; 274(50): 36009–36014: (1999).
CrossRef - Babgohari M. Z, Ebrahimie E, Niazi A. In silico analysis of high affinity potassium transporter (HKT) isoforms in different plants. Aquatic Biosystems; 10: 9: (2014).
CrossRef - Yin X, Zhao Y, Luo D, Zhang H. Isolating the promoter of a stress-induced gene encoding betaine aldehyde dehydrogenase from the halophyte Atriplex centralasiatica Iljin. Biochimica et Biophysica Acta (BBA) – Gene Structure and Expression; 1577: 452–456: (2002).
CrossRef - Pandey G. K. Eds. Elucidation of abiotic stress signalling in plants: Functional genomics perspectives. Springer 2; (2015).
- Ciolkowski I, Dierk W, Birkenbihl R. P, et al. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Molecular Biology; 68(1-2): 81–92: (2008).
CrossRef - Tran L. S. P, Nakashima K, Sakuma Y, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell; 16: 2481-2498: (2004).
CrossRef - Gao F, Xiong A, Peng R, et al. OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell, Tissue and Organ Culture; 100: 255–262: (2010).
CrossRef - Meier I, Gruissem W. Novel conserved sequence motifs in plant G-box binding proteins and implications for interactive domains. Nucleic Acids Research; 22(3): 470–478: (1994).
CrossRef - de Jager S. M, Menges M, Bauer U. M, et al. Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Molecular Biology; 47: 555-568: (2001).
CrossRef

