Introduction
Tomatoes (Solanum lycopersicum) are considered one of the most extensively cultivated crops globally, contributing significantly to agricultural output and economic value because of its demand throughout the year. Food and Agriculture Organization1 (FAO) data indicates that global tomato yield exceeds 180 million tons per annum, with India occupying the position of second largest producer. Notwithstanding their economic significance, tomato plants exhibit susceptibility to diseases that can substantially impact yields, with annual losses estimated between 20% and 40%.
Conventional approaches to disease detection, such as visual assessment or laboratory analysis, necessitate skilled professionals and considerable time. These methodologies are also susceptible to human error due to environmental variations and the similar symptomatology exhibited by different diseases2. In rural or resource limited regions, the paucity of experts and diagnostic facilities further complicates timely disease management.
The revolutionary Machine Learning (ML) and Deep Learning (DL) has transformed agricultural practices3, particularly in automating plant disease detection and classification. Among these techniques, Convolutional Neural Networks (CNNs) are particularly noteworthy for their capacity to learn hierarchical features directly from image data, obviating the need for manual feature extraction. However, while DL models excel under controlled conditions, their practical implementation faces challenges, including data diversity and scalability. This objective of this paper is to provide a comprehensive examination of tomato disease detection techniques, with a focus on CNN based models, their efficacy, and approaches to overcome real-world obstacles. CNNs breaks down images into hierarchical layers of features and outperform traditional methods like manual feature extraction, rely on predefined rules or human expertise to detect specific features (e.g., leaf spots or discoloration).
Tomato disease detection has garnered significant attention due to its impact on agricultural productivity. Recent advancements leverage deep learning and image processing techniques to enhance detection accuracy and efficiency. Various methodologies have been proposed4, each addressing specific challenges in identifying diseases in tomato plants. Several studies utilize CNNs for automated disease detection, effectively learning complex patterns from tomato leaf images. Parallel CNN5 approach has also used in literature to reduce the training time. For instance, a CNN-based model achieved over 90% accuracy in classifying diseases like leaf mold and target spot (Sonawane, 2023)(R., 2024).During disease detection6,7 , researcher has also emphasis on the feature extraction techniques which incorporate self-attention mechanisms and dynamic activation functions to improve feature extraction, resulting in a mean Average Precision (mAP).Many researcher has used image processing methods8,9 to enhance visual features of tomato plants, allowing for precise identification of diseases like early and late blight. This approach aids in distinguishing between various disease types through unique feature signatures. Bootstrapping approach10 and deep CNN11,12 are extensively used in agriculture domain for identification of diseases for different plants13.
Materials and Methods
The study utilized a combination of publicly available and custom collected datasets like Plant Village Dataset and real world dataset as refered in Table 1. Plant village dataset contains over 14,500 annotated images of tomato leaves, representing both healthy and diseased samples. It includes classes such as bacterial spot, early blight, late blight, and septoria leaf spot.
RealWorld dataset contains 5000 field images which were collected under varying environmental conditions to simulate real-world diversity. These images featured heterogeneous lighting, complex backgrounds, and varied resolutions. Table1 shows the brief summary of dataset used under consideration.
Table 1: Dataset Information
| Dataset | Number of Images | Classes | Conditions |
| Plant Village Dataset | 14,500 | 10 | Controlled |
| RealWorld Dataset | 5,000 | 6 | Uncontrolled |
In this paper, the advanced techniques for tomato disease detection which makes use of machine learning and deep learning approaches are explained. Preprocessing is critical for ensuring model robustness. During preprocessing resizing, normalization and augmentation are carried out on dataset. All images were standardized to 224×224×3 pixels to align with the input requirements of convolutional neural network14 architectures. Pixel values were scaled to the range [0, 1] to enhance computational efficiency. Techniques such as cropping, rotation, flipping and zooming are applied at data augmentation stage to artificially expand the dataset and address class imbalances. The main goal of data augmentation is to increase the diversity of the training data important when the dataset is limited or have imbalanced distribution, as it helps reduce the risk of overfitting and allows the model to learn more robust and varied patterns. In this paper three different approaches are applied and tested as explained in following section.
Approach1: DenseNet
The model combines a pre-trained DenseNet201 architecture with a CNN classifier, chosen for its superior accuracy compared to other models. DenseNet architecture15 is applied here to achieved the best accuracy due to its densely connected layers that optimize feature reuse. DenseNet201 extracts features which are processed by the convolutional neural network for classification. The model’s performance is evaluated using test and validation sets. The initial stage involves data pre-processing, followed by data augmentation as second stage.
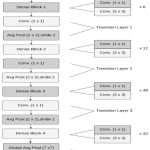 |
Figure 1: DenseNet Model |
The third stage introduced the DenseNet201 that leverages transfer learning. DenseNet 201 without human intervention extract features and apply the weights pre-trained on the ImageNet dataset, thereby reducing computational demands. DenseNet201’s structure enables the development of uncomplicated models. It also allows for feature reuse across layers, enhancing parameter efficiency and enabling greater diversity in successive layers, thus improving performance. In a feed forward approach every layer is connected to successive layer and creates a direct pathway for information to flow throughout the network. Moreover, DenseNet201 incorporates a pooling layer and bottleneck structure, which minimizes model complexity and parameter count, increasing efficiency. Every layer in the DenseNet201 network performs a nonlinear transformation to capture complex patterns. Each layer comprising of convolution (Conv), pooling, rectified linear units (ReLU), and batch normalization (BN). DenseNet201 employs a unique approach where all layers are connected in such as way that the output of one layer is used as input of successive layers. This dense connectivity pattern results in a total of N(N+1)/2 connections in an N-layer network, significantly increasing the flow of information and improving the model’s ability to reuse features from earlier layers. In this paper, the DenseNet201 structure consists of more than 700 layers and more than 20 million parameters. The input layer is configured to accept images with dimensions of 224 x 224 x 3. The DenseNet201 structure is illustrated in Fig.1.
During the fourth stage, the classification layers are eliminated, and six new layers are introduced for the classification task. The initial layer and second layer consists of 1024 neurons and 512 neuron respectively. Both layers utilizing a ReLu activation function. A dropout layer is implemented as the third layer, with a dropout rate of 0.2 to mitigate the effect of overfitting. The fourth layer incorporates a global average pooling layer for reduction in feature maps. The fifth layer is comprising 128 neurons and ReLu function. A second dropout layer is also introduced with a 0.2 dropout rate serves as the sixth layer. The final layer consists of only 10 neurons and a Softmax function, which outputs the 10 classes of tomato diseases. The subsequent section provides a detailed examination of the proposed model’s results and compares them to other models.
Approach 2: ResNet50
In place of DenseNet, ResNet is implemented which is known for its residual learning framework, which mitigates the vanishing gradient problem. First two phases are remain same i.e. preprocessing and augmentation where after data collection, a cleaning process is implemented to remove faulty images from the dataset. Images are resized to 224 × 224 pixels, which optimizes machine load during training and yields optimal results. The images are then labeled using a one-hot encoding system and converted into a NumPy array for faster computation. ResNet has several variants based on CNN principles but with varying layer counts. In the third phase, a deep ResNet5016 model which comprises of convolutional layers(CONV), identity block(IB), convolutional block (Conv Block), and the fully connected as shown in fig. 2. ResNet50 refers to the 50-layer neural network variant where convolutional layers are responsible for extracts various features from input images while the IB and Conv block are responsible for processing and transforming these features to train the model. The final phase utilizes a fully connected layer for classification. The accuracy of a deep CNN model is heavily influenced by dataset quality.
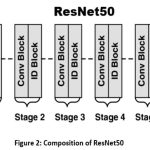 |
Figure 2: Composition of ResNet50 |
Approach 3: VGG16
VGG-16 is a widely adopted CNN architecture acronym as “VGG” stands for Visual Geometry Group. VGG architecture17 is particularly renowned for its effectiveness with ImageNet, an extensive project used for visual object recognition. This model is highly regarded in the field of image classification within deep learning due to its exceptional utility. In VGG16 “16” indicates the number of layers in the neural network which comprises of 13 convolutional Layers, 5 pooling layer and 3 dense layers as shown in fig. 3.
 |
Figure 3: Composition of VGG 16 |
Experimental Results
The three models were evaluated using the following metrics:
Accuracy: Percentage of correctly classified samples.
Precision, Recall, F1Score: Metrics for class wise performance analysis.
Confusion Matrix: Visualized misclassifications across disease classes.
DenseNet achieved the highest overall accuracy of 99.9% under controlled conditions, significantly outperforming other architectures as shown in table 2. However, when tested on real-world datasets, all models exhibited a performance drop, highlighting challenges in generalization. Models may not generalize well to diverse environments, leading to reduced accuracy and robustness.
Table 2: Model Performance Accuracy
| Controlled Environment | Real World Environment | Drop (%) | |||||||||
| Model | TP | FP | FN | TN | Acc. (%) | TP | FP | FN | TN | Acc. (%) | |
| DenseNet | 13997 | 11 | 10 | 482 | 99.9 | 2132 | 866 | 760 | 1242 | 67.5 | 32.4 |
| ResNet50 | 13685 | 170 | 190 | 455 | 97.6 | 2105 | 1003 | 783 | 1109 | 64.3 | 33.3 |
| VGG16 | 13385 | 340 | 465 | 310 | 94.5 | 2002 | 1106 | 883 | 1009 | 60.2 | 34.3 |
While comparing the training time of these models VGG16 takes less training time as compare to other two approaches as shown in table 3.
Table 3: Model Training Time
| Model | Training Time (hrs.) | Parameter Count (millions) | Notable Features |
| DenseNet | 8.5 | 28 | Dense connections |
| ResNet50 | 7.2 | 23 | Residual blocks |
| VGG16 | 6.8 | 138 | Sequential architecture |
Discussion
A common deep learning issue which was observed is increase in training and test error rates as layer count increase. Among CNN architectures18,19 particularly DenseNet and ResNet, excelled in capturing complex features, reducing the need for manual preprocessing. Their scalability and adaptability to different image datasets make them suitable for agricultural applications. Some of the challenges associated with CNN architecture are overreliance on controlled datasets hampers real-world performance which results in dataset bias, Training CNNs requires significant computational resources like GPU etc. and Real-time processing on low power devices remains a challenge during field deployment. As deep learning is evolving need to have advance techniques for improvements to the current methodology especially for real time validation and suggest areas for further investigation, such as expanding the dataset or exploring new feature extraction techniques.
Conclusion
This paper underscores the potential of deep learning in automating tomato disease detection. In this paper three different approaches of deep learning are applied and tested on controlled environment and realtime environment. DenseNet,ResNet50 and VGG16 demonstrated near perfect accuracies in controlled settings, though significant challenges persist in adapting these models for real-world use. While the advancements in CNN models for tomato disease detection are promising, challenges remain in achieving consistent performance across varying environmental conditions and image qualities. Further research is needed to enhance model robustness and reduce false positives in complex agricultural settings. Future work should focus on expanding datasets to include diverse environmental conditions, developing lightweight models for mobile and IoT devices, enhancing interpretability to build farmer centric solutions. By addressing these gaps, deep learning technologies can revolutionize agricultural disease management, improving productivity and ensuring global food security.
Acknowledgement
We extend our sincere thanks to everyone who directly or indirectly played a role in this work. A special appreciation goes to AISSMS Institute of Information Technology for providing experimental setup environment.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest
Data Availability Statement
The manuscript incorporates all datasets produced or examined throughout this research study.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
As part of our commitment to ethical research practices, all participants involved in data collection for this project provided informed consent.
Author Contributions
Meenakshi Thalor: Initiated the research work by outlining, mentioning objectives and prepared the system architecture. Contributed in customized data collection using camera and in documentation of paper.
Yash Chavhan: Implementation of system starting from data collection to classification task.
Sanjay Mate: Performed the validation of model by using different evaluation measures like precision, recall and f score.
References
- Islam M.P., Hatou K., Aihara T., Seno S., Kirino S., Okamoto S. Performance prediction of tomato leaf disease by a series of parallel convolutional neural networks. Smart Agricultural Technology. 2022:2:100054. https://doi.org/10.1016/j.atech.2022.100054.
CrossRef - Bouni, Mohamed & Hssina, Badr & Khadija, Douzi & Douzi, Samira. Impact of Pretrained Deep Neural Networks for Tomato Leaf Disease Prediction. Journal of Electrical and Computer Engineering. 2023:1-11. 10.1155/2023/5051005.
CrossRef - Sudar, K. Muthamil & Nagaraj, P. & Prakash, Bhanu & Reddy, M. & Naidu, M. & Kumar, Hemanth. Development of Tomato Leaf Disease Prediction System to the Farmers by using Artificial Intelligent Network. 6th International Conference on Intelligent Computing and Control Systems (ICICCS) 2022:955-961. 10.1109/ICICCS53718.2022.9788189.
CrossRef - Verma S., Chug A. & Singh A. Prediction Models for Identification and Diagnosis of Tomato Plant Diseases. International Conference on Advances in Computing, Communications and Informatics (ICACCI), 2018:1557-1563. 10.1109/ICACCI.2018.8554842..
CrossRef - Pacal, I., Kunduracioglu, I., Alma, M.H. et al. A systematic review of deep learning techniques for plant diseases. Artif Intell Rev. 2025: 57:304. https://doi.org/10.1007/s10462-024-10944-7
CrossRef - Barth R., Jsselmuiden J., Hemming J., Van Henten E.J.. Synthetic bootstrapping of convolutional neural networks for semantic plant part segmentation, Computers and Electronics in Agriculture. 2012:16:291-304 . https://doi.org/10.1016/j.compag.2017.11.040.
CrossRef - Lu, J., Tan, L., & Jiang, H. Review on Convolutional Neural Network (CNN) Applied to Plant Leaf Disease Classification. Agriculture, 2021:11(8):707. https://doi.org/10.3390/agriculture11080707
CrossRef - Mohanty SP, Hughes DP and Salathé M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016: 7:1419. doi: 10.3389/fpls.2016.01419
CrossRef - Zhang Y-D, Dong Z, Chen X. Jia W, Du S, Muhammad K, et al. Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed Tools Appl . 2019:78(3):3613-32
CrossRef - Wu Y, Xu L, Goodman ED. Tomato Leaf Disease Identification and Detection Based on Deep Convolutional Neural Network. Intelligent Automatic Soft Computing.2021; 28;561-576.
CrossRef - Afifi, A., Alhumam, A., & Abdelwahab, A. Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Plants. 2021;10(1); 28.
CrossRef - Ji, M., Zhang, L., & Wu, Q. (2020). Automatic grape leaf diseases identification via UnitedModel based on multiple convolutional neural networks. Information Processing in Agriculture, 2020;3; 418–426.
CrossRef - Liu J. and Wang X., Plant diseases and pests detection based on deep learning: A review,Plant Methods, 2021:17.
CrossRef - Su T., Li W., Wang P., and Ma C. Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Frontiers in Plant Science, 2019:10:705.
CrossRef - Huang G., Liu Z., Van Der Maaten L. and Weinberger K.Q. Densely Connected Convolutional Networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017: 2261-2269. doi: 10.1109/CVPR.2017.243.
CrossRef - Tian X.and Chen C. Modulation Pattern Recognition Based on Resnet50 Neural Network, IEEE 2nd International Conference on Information Communication and Signal Processing (ICICSP). 2019:34-38. doi: 10.1109/ICICSP48821.2019.8958555.
CrossRef - Qassim H., Verma A. and Feinzimer D. Compressed residual-VGG16 CNN model for big data places image recognition, IEEE 8th Annual Computing and Communication Workshop and Conference (CCWC). 2018:169-175. doi: 10.1109/CCWC.2018.8301729.
CrossRef - Chaudhary, A., & Raheja, N. Exploration of Tomato Leaves Diseases Classification Through Advanced Machine Learning and Deep Learning Models. In 2024 International Conference on Cybernation and Computation (CYBERCOM).2024:742-746.
CrossRef - Singh, A., Kumar, S., & Choudhury, D. ,Tomato Leaf Disease Prediction Based on Deep Learning Techniques. In International Conference on Computation of Artificial Intelligence & Machine Learning.2024:357-375
CrossRef

