Introduction
Soil serves as an ideal dwelling place for numerous kinds of microorganisms including bacteria, fungi, archaea, and protozoa, apart from being an abundant habitat for diverse organisms.1 Many microorganisms that inhabit the soil are necessary for the survival of life on earth. The significance of microorganisms has grown due to their ability to manufacture industrially valuable enzymes.2 The discovery of extracellular enzymatic activity in many microbes has drawn significant attention in recent years due to the potential of employing these organisms as biotechnological sources of industrially relevant enzymes.3 The main class of soil microorganisms are bacteria, which are distinguished by their ability to create a wide variety of extracellular enzymes including amylase, cellulase, protease, lipase, pectinase and chitinase. According to Morris and Blackwood,4 bacteria can have up to 10 billion cells/g in soil and are engaged in the breakdown of organic materials, nutrient cycling, soil aggregation and humus formation. In modern biotechnology, one of the most commonly used enzymes is starch hydrolytic amylase. Due to the abundance of starchy compounds found in soil, it is one of the richest sources of amylases.2 The enzyme amylase breaks down carbohydrate molecules into smaller products. Its main substrate is starch, which is broken down into smaller components such as dextrin, maltose and glucose. The enzyme basically hydrolyzes the α-1,4 glycosidic bonds that hold the glucose units together.5 The amylase enzyme has a wide range of applications in a number of industrial processes such as food, beverage, fermentation, paper, textiles and pharmaceutical industries.6,7 The widespread use of amylase has increased awareness of the rise in domestic amylase production. Recent developments in biotechnology have led to greater use of the amylase enzyme in clinical research and medicine8.
The quantities of minerals and nutrients in different types of soils vary. As a result, numerous reports on microorganisms from various soil types that degrade starch have been published. Due to their short growing period, low cost of production, eco-friendly nature, minimal worker handling concerns, productivity and ease of bacterial gene editing, enzymes from microbes are an ideal choice for industrial production.9 In order to grow and maintain their metabolic processes, microorganisms need a few fundamental nutrients.10 Microorganisms ability to synthesize amylase is dependent on the strain type, growth medium, cultivation technique, cell growth, nutritional needs, incubation time, pH, temperature, metal particles, and thermostability.11
Pseudomonas spp., Bacillus licheniformis and Bacillus amyloliquefaciens are the most commonly used bacterial species for the extraction of enzymes. The process of producing the enzyme amylase is also detected in other species, such as Bacillus subtilis and Bacillus cereus. Amylases derived from Bacillus licheniformis, Bacillus stearothermophilus, and Bacillus amyloliquefaciens showed potential in an array of commercial applications, including fermentation, food processing, textile and paper industries.12 Many amylase-producing bacteria have been identified to date, however no research has been published on amylase producing bacteria from soil collected in the Biligiriranga hills, Karnataka. The microbial biodiversity in this area appears to be abundant, which may be helpful for producing industrial enzymes like amylase. This research study is the first to find amylase producing bacterial isolates from soil sample of Biligiriranga hills. In the future, these bacterial amylases can be used locally in a variety of industries such as agriculture, animal feed, bio ethanol, food, textile, brewing, pulp and paper.
Materials and Methods
Collection of soil sample
Soil samples were collected from the south Indian hill ranges known as the Biligiriranga Hills, which are located in south-western Karnataka near the Tamil Nadu border (11.1271° N, 78.6569° E). Using a sterile spatula, soil samples were removed from the rhizospheres and placed inside the sterilized bag before being brought to the lab for further processing.
Isolation of bacteria
Isolation of bacteria from soil samples
To isolate the bacteria, collected soil samples were homogenized and subjected to the serial dilution method. In brief, one gram of soil sample was separately transferred into 9 ml of distilled water containing 1 % of peptone and mixed well for at least 1 min. This suspension was serially diluted from 10-1 to 10-3 and accordingly, 0.1ml solution was taken from 10-3 suspension and spread out on Starch agar (5.0 g/L of peptone, 5.0 g/L of sodium chloride, 1.5 g/L of yeast extract, 2 g/L of soluble starch and 1.5 gL of agar ; pH 7.4) plates. The inoculated plates were incubated at 37° C for 24h until typical bacterial colonies were obtained.13
Preparation of pure culture
Following incubation, the bacterial colonies showing a clear difference in morphology were picked up from the plates and sub cultured on NA slants by streaking method. The slants were incubated at 37° C for 24h to get a pure culture of bacterial isolate and then preserved at 4° C in the refrigerator until further studies.14
Screening and selecting amylase producing bacteria (starch iodine test)
The capacity of a microbe to break down starch is a criterion for determining its production of amylase. In this study purified bacterial colonies were examined for amylase production following the standard protocol.15 The bacterial colonies were inoculated in freshly prepared starch agar plates using a sterile inoculation needle by dot method and incubated at 37° C for 24-48h. Following the incubation period, the culture plates were flooded with Gram’s iodine solution, and the plates were kept undisturbed for 5–10 minutes. Then, the iodine solution was carefully decanted from the plates, and any clean zone development surrounding the colonies was observed for a positive result. The isolates that showed clear zones around them were chosen for further studies.
Identification and characterization of bacteria
Morphological and Biochemical Characterization
The selected bacterial isolates that produced amylase were identified by both microscopic and macroscopic examination. Gram staining was done to ascertain the morphological characterization of the isolated bacteria. It is a crucial microbiological technique used to identify organisms, where methylene blue and crystal violet are used as the primary colours for staining. Gram-positive organisms retain the primary colour and appear purple under a microscope, while Gram-negative organisms appear red. Following Gram’s reagent staining, the isolates were observed under a light microscope to classify the bacteria as Gram negative or positive based on the colour and shape of the colony.16 According to Bergey’s Manual, 17 different biochemical tests such as catalase, potassium hydroxide and methyl red tests were carried out to characterize bacterial isolates.
Catalase Test
The purpose of the test is to determine whether a bacterium produces the catalase enzyme. The presence of catalase enables the breakdown of hydrogen peroxide into liquid water (H2O) and oxygen gas (O2), which results in the formation of bubbles, signifying a successful reaction. In this test, bacterial culture was mixed with a drop of hydrogen peroxide on a glass slide and observed for bubble formation.18
Potassium hydroxide (KOH) test: This test could be useful in distinguishing between organisms that are Gram positive or negative. Gram negative bacteria have thin peptidoglycan-coated cell walls that are dissolved by KOH. When cell lyses, the mixture becomes viscous or stringy due to the released cellular DNA, which results the formation of mucous thread within 30 seconds. However, KOH has no effect on the cells of Gram-positive bacteria. In order to perform this test, the bacterial culture was mixed with 2-3 drops of 3% KOH on a glass slide using a tooth pick and observed for the mucoid string formation by lifting the toothpick.19
Methyl Red Test
The methyl red (MR) test determines whether enough acid is produced by the bacterial cells using glucose present in the MR media. The bacterial culture was inoculated in 10 ml Methyl Red–Voges–Proskauer broth (containing 0.05 g/ml of glucose, 0.07 g/ml of peptone and 0.05 g/ml of potassium phosphate with the pH indicator methyl red) and incubated at 37° C for 48 h. Five drops of the methyl red indicator was added to the broth followed by incubation, the tubes were shaken well and were allowed to stand for 30 min to 1 h. A change in the broth colour from yellow to red was observed, where the glucose is utilized by the bacteria to produce a stable acid.20
Molecular evaluation of bacterial isolate
DNA isolation of bacteria
For the molecular identification of the strain producing amylase enzyme, the genomic DNA was extracted from 24 h bacterial culture by Phenol-Chloroform extraction method. The DNA quality was visualized under UV light and quantified spectrophoto-metrically at OD 260/280 nm and then used for polymerase chain reaction (PCR) analysis.
PCR amplification of 16S rRNA gene
Amplification of the 16S rRNA gene was performed using the universal primer set of BSF-5′-GAGTTTGATCCTGGCTCAGG-3′ and BSR-5′-TCATCTGTCGTCCCACCTTCGGC-3′. The primers were designed using sequence with accession number MT020289.1. The PCR reactions were conducted in 25 μl volumes with 2 μl of the genomic DNA sample, 0.75 U Taq DNA polymerase (Takara, Japan), 2.5 X PCR buffer with 2.5 mM MgCl2, 0.2 mM dNTP Mix and 20 pmol of F/R primers. Amplification was carried out in a thermal cycler (Applied Biosystems) with initial denaturation at 95° C / 4 min, followed by 35 cycles (95° C for 30 sec, 56° C for 1 min and 72 ° C for 30 sec) and a final extension for 72° C/10min.
Agarose gel electrophoresis
The PCR products were then electrophoresed in 1.2 % agarose gel with ethidium bromide and exposed to ultraviolet (UV) light for visualization in Gel doc/UV transilluminator (Biobee Transilluminator, Bangalore, India).
Sequencing of 16S rRNA and construction of Phylogenetic tree
PCR products were subjected to DNA sequencing by the Taq dye-deoxy terminator method (DNA sequencer, Applied Biosystems). The obtained 16S rRNA gene sequence was Blast searched (http://www.ncbi.nlm.nih.gov/) and aligned with most similar bacterial species found in NCBI nucleotide database. The bacterial isolate was assigned to a genus based on 99–100 % similarity index. The Phylogenetic tree was constructed using the packages DNADIST and Bootstrap.
Evaluation of growth conditions for bacterial growth
Effect of pH on bacterial growth
In order to investigate the effect of pH on bacterial growth, 50 ml of starch-nutrient broth with different pH values (6, 7, 8 and 9) was poured into four separate Erlenmeyer flasks. Each flask was inoculated with the bacterial culture (Sample 2) and incubated at 37° C for 24h. After 24h, the optical density (OD) at 600 nm was recorded for each broth with a varied pH.
Effect of temperature on bacterial growth
To investigate the effects of temperature on the growth of bacterial cells, 50 ml of starch-nutrient broth was added to 4 separate flasks and inoculated with the bacterial culture (Sample 2). The inoculated cultures were incubated for 24h at different temperatures like 30° C, 37° C, 40° C and 45° C, in the incubators. The absorbance of the broth incubated at each temperature was determined at 600 nm.
Effect of incubation period on bacterial growth
To investigate the effect of incubation time on the amylase-producing bacteria (Sample 2), the growth of the bacteria was determined by adding bacterial culture to a flask containing 50 ml of starch-nutrient broth. The experiment was conducted by incubating the bacteria at different incubation period, such as 24, 36, 48, and 72h. The growth of the bacteria was checked at each incubation time by measuring the OD at 600 nm.
Statistical Analysis
The data was determined with three independent replicates, and the data were the mean of three parallel experiments.
Results
Isolation and purification of bacterial isolates from soil sample:
In this study, soil samples from the B.R. Hills (11.9988° N, 77.1398° E) were serially diluted to isolate bacteria, spread on a starch agar medium, and incubated at 37 °C for 24 hours. A dense, thick population of mixed bacterial colonies originating from a dilution factor of 10-3 was observed on the starch agar plates (Fig. 1). Pure culture of 15 bacterial isolates (Sample 1 to 15) were obtained on fresh NA slants (Fig. 2), which were randomly picked up from the mass bacterial population based on their colony morphology.
Screening and selecting amylase producing bacteria
Fifteen pure bacterial isolates were evaluated for amylolytic activities using starch agar medium, out of which sample 1, 2, and 3 gave a distinct clear zone around the bacterial culture indicating a positive result for starch hydrolysis. The presence of clear zones around bacterial growth suggested that the bacteria were able to hydrolyze the starch, creating the starch-free clear zone. Especially the areas around the bacterial growth, was where it occurred, while the remaining portion of the plate still contained non-hydrolyzed starch with dark blue/purple colour indication (Fig. 3).
Morphological and Biochemical Identification of amylase producing bacteria
Isolated bacterial colonies were microscopically and macroscopically observed. The organisms retained the primary colour and appeared purple-blue and rod shaped under the microscope (Fig. 4). Various biochemical tests including catalase, potassium hydroxide and methyl red tests were done to identify bacteria. The biochemical characteristics of the isolates are summarized in Table 1. The isolates showed bubble formation when mixed with hydrogen peroxide indicated positive for catalase test (Fig. 5). When the bacterial culture was mixed with potassium hydroxide, no mucoid threads were formed. This showed that, the KOH test results for all three isolates were negative (Fig. 6). In Methyl Red test, the broth colour was remained unchanged, which indicated the tested isolates were negative for this test (Fig. 7). Based on morphological characteristics, microscopic and biochemical tests the isolates were Gram positive isolates.
Molecular characterization based on 16s rRNA
Molecular identification using 16S rRNA sequencing confirmed the selected isolate (Sample 2). Agarose gel electrophoresis was used to do a qualitative examination of the isolated DNA and to evaluate the existence of pure DNA. The isolated genomic DNA was tested for purity on the agarose gel and a clear band was seen, proving its purity (Fig. 8A). The PCR product was analysed on the agarose gel to verify the amplicon size (1200 bp) corresponding to the strain (Fig. 8B).
The BLAST analysis of the obtained nucleotide sequence showed maximum homology with the earlier NCBI database. 16S rRNA gene sequence of bacterial isolate strain B showed 100 % homology with earlier NCBI submissions of Bacillus subtilis 16S rRNA gene sequences (Accession number: MT020289.1, OP900551.1, MW365355.1). This molecular information was supporting with the morphological and biochemical data. The Phylogenetic tree also confirmed significant relation with B. subtilis. The similarity between the 16S rRNA gene sequences of B. subtilis and strain B is depicted in dendrogram (Fig. 9). Thus, on the basis of morphology, biochemical and molecular analysis the bacterial isolate was confirmed as B. subtilis. The 16S rRNA partial sequence has been deposited in the NCBI GenBank as B. subtilis Sample 2 with the accession no. PP112150 (Fig. 10).
Evaluation of growth conditions for bacterial growth
Effect of pH, temperature and incubation period on bacteria growth
An ideal pH and temperature is required by the bacterial cell to carry out its molecular and metabolic processes. Different bacterial species have different optimal pH and temperature values. To check the optimum conditions the bacterial isolates were inoculated in starch medium with different pH, temperature and incubation periods. After incubation, the OD at 600 nm was measured for each culture grown at different pH, temperatures, and incubation times. The absorbance of each test was recorded and presented in a bar graph (Figs. 11, 12, and 13). The optimum pH for B. subtilis growth was found at pH 7, with significantly lower bacterial growth observed at pH 6, 8, and 9. Similarly, bacterial growth was found to be highest at 37° C and not at other temperatures such as 30° C, 40° C, and 45° C. The incubation time at which bacterial growth was highest was 36h, while lower bacterial growth was observed at 24, 48, and 73h. These results suggest that 37° C, pH 7, and 36 h of incubation were the ideal parameters for B. subtilis.
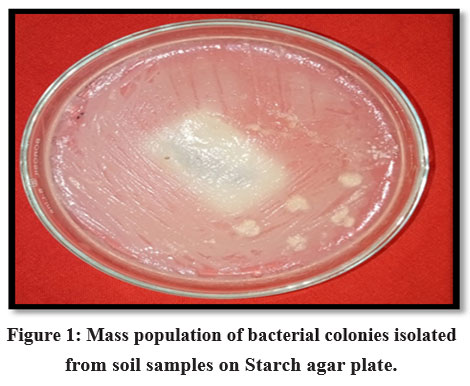 |
Figure 1: Mass population of bacterial colonies isolated from soil samples on Starch agar plate. |
 |
Figure 2: Pure culture of bacterial isolates on Nutrient agar slants. |
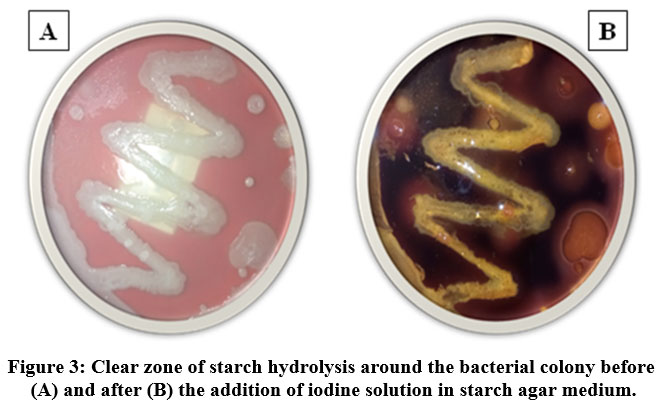 |
Figure 3: Clear zone of starch hydrolysis around the bacterial colony before (A) and after (B) the addition of iodine solution in starch agar medium. |
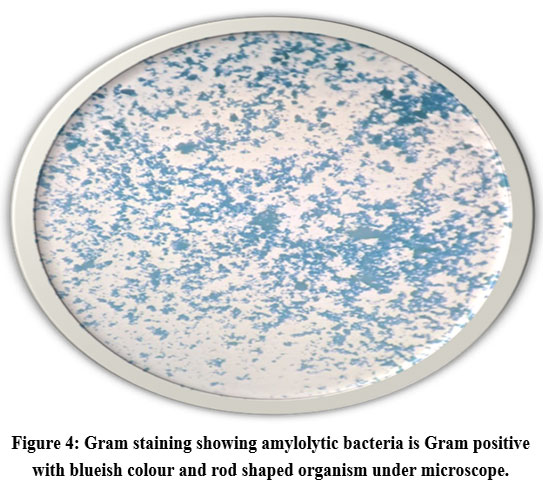 |
Figure 4: Gram staining showing amylolytic bacteria is Gram positive with blueish colour and rod shaped organism under microscope. |
 |
Figure 5: Catalase Test: Amylolytic bacteria showing bubble formation as a positive result of oxygen release by hydrogen peroxide breakdown . |
 |
Figure 6: Potassium hydroxide solubility test: No mucoid thread formation, indicates that the amylolytic bacteria are gram positive. |
 |
Figure 7: Methyl red test: No red colour change in MR broth after adding the pH indicator methylene red, representing negative result for the bacterial isolates. |
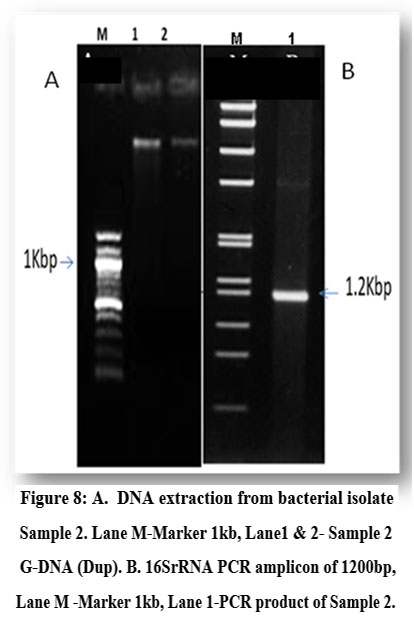 |
Figure 8: A. DNA extraction from bacterial isolate Sample 2. Lane M-Marker 1kb, Lane1 & 2- Sample 2 G-DNA (Dup). B. 16SrRNA PCR amplicon of 1200bp, Lane M -Marker 1kb, Lane 1-PCR product of Sample 2. |
 |
Figure 9: Dendrogram illustrating the similarity of nucleotide sequences of the 16S rRNA genes of strain B to that of Bacillus subtilis. Phylogenetic tree of Bacillus strains based on the nucleotide sequences of the partial 16S rRNA genes. |
 |
Figure 10: Bacillus subtilis strain B 16S ribosomal RNA gene, partial sequence (Accession no. PP112150). |
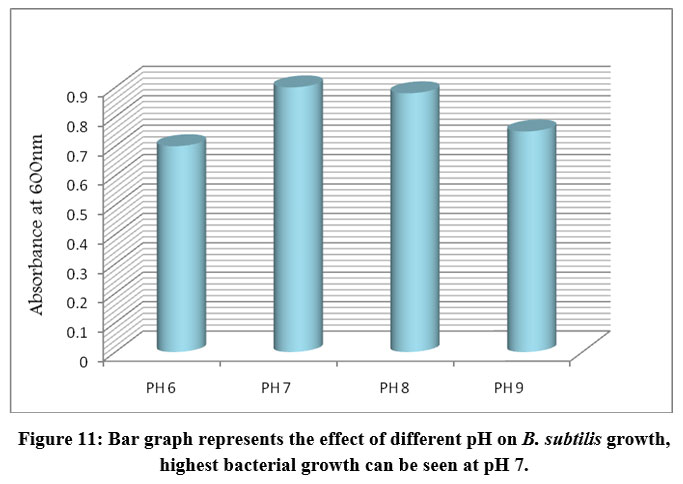 |
Figure 11: Bar graph represents the effect of different pH on B. subtilis growth, highest bacterial growth can be seen at pH 7. |
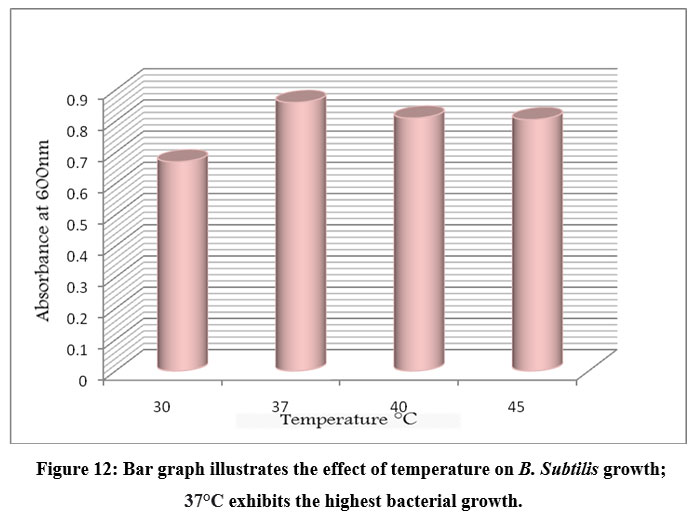 |
Figure 12: Bar graph illustrates the effect of temperature on B. Subtilis growth; 37°C exhibits the highest bacterial growth. |
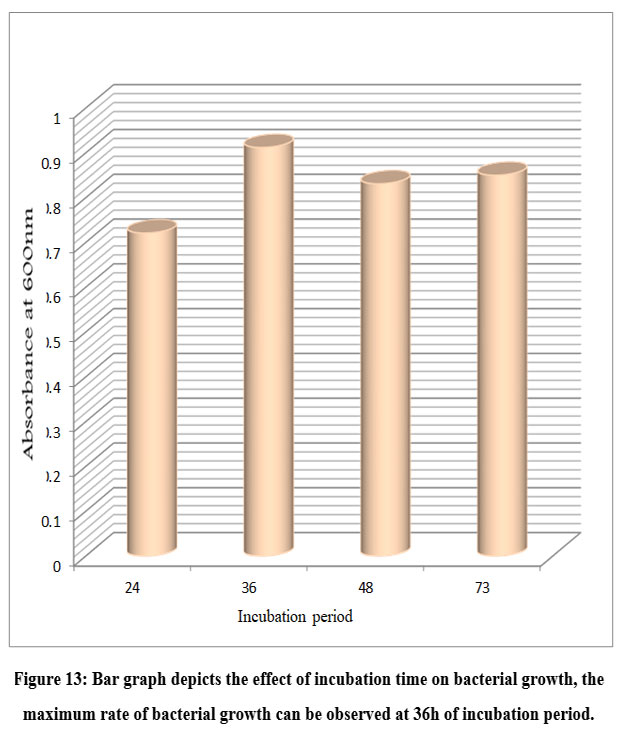 |
Figure 13: Bar graph depicts the effect of incubation time on bacterial growth, the maximum rate of bacterial growth can be observed at 36h of incubation period. |
Discussion
Soil contains a vast array of microorganisms that generate a wide range of enzymes. Enzymes are the proteins produced by living things in biological systems that catalyze chemical reactions.1 Among these are amylases, which hydrolyze starch into its smaller monomer components, such as glucose. With these characteristics, amylase an enzyme produced by a variety of bacteria is a widely utilized enzyme with many industrial uses. Amylase-producing microorganisms can be isolated from a variety of environments, such as farms, water sources, food processing plants, and so on. On the other hand, soil is known to be the main source of amylase-producing microorganisms and serves as their repository.
In the current study, amylase-producing bacteria were isolated from the soil source collected from BR Hills, Karnataka; the isolated bacteria were predominantly screened for amylase production on starch agar medium. The best amylase-producing bacteria were those with higher clear zone formation. Out of 15 Sample 1, 2 and 3 were identified as best amylase producer and were characterized both morphologically and biochemically. The bacteria were confirmed as gram-positive and rod-shaped under a microscope observation. Biochemically the isolates indicated positive for the catalase test, however negative for the potassium hydroxide test and methyl red test. Based on the above results and according to Bergey’s manual, the isolates were identified as Bacillus spp. The amylase-producing isolate strain B upon molecular analysis, 16S rRNA sequencing results and Phylogenetic tree confirmed it as B. subtilis.
Our identification was in accordance with earlier reports, where many scientists have isolated amylase-producing bacteria belongs to distinct species from soil samples, performed biochemical experiments and validated their authenticity through 16S rRNA sequencing. The current finding is consistent with the finding of Namo et al.10 who isolated and characterized the six amylase producing bacteria from soil of Debre Berhan city, Ethiopia and identified as Bacillus spp. Silaban et al. 21 isolated and identified the amylolytic bacteria from rice soil samples, while Nimisha et al. 22 isolated from garden soil. Likewise, the amylase producing B. subtilis was isolated from soil samples, confirmed its identification through different biochemical tests and 16S rRNA sequence.23, 24 Bacillus species isolated from the soil of potato dump sites in Madhya Pradesh were identified based on morphological and biochemical tests.25 Ghazala et al.26 and Srivathsan et al.11 conducted a study on the screening and isolation of amylase-producing bacteria from soil samples, identifying them as Bacillus sp. based on 16S rRNA sequencing. Similar results were supported by Singh and Kumari27, where they isolated, characterized the amylase producing Bacillus sp. from soil samples of Uttarakhand, India and Luang-In et al.28 identified twelve amylase-producing Bacillus spp. from the unexplored Nasinuan Forest, Thailand, using 16S rRNA sequencing. Comparatively, Bacillus cereus KR9 was found as a putative amylolytic bacterial isolate by Krishma and Radhathirumalaiarasu 29 in another study. Six amylase-producing species were reported from garbage and garden soil, which included Bacillus azotoformans, B. stearothermophilus, B. acidocaldarius, B. subtili, B. megaterium, and Acetobacter liquefaciens. 2 Rakaz et al. 30 identified active α-amylase producers in Khartoum State, Sudan, as Bacillus cereus and Bacillus licheniformis through biochemical characterization and 16S-rDNA sequencing. Furthermore, amylase-producing Pseudomonas aeruginosa from soil samples in Tuljapur, was identified through morphology, biochemical tests and 16s rRNA sequence.31 Two amylase producing bacterial isolates were isolated from sugarcane and areca plant soil samples, identified based on biochemical tests such as indole, nitrate, methyl red, and Catalase.32
One of the critical factors that significantly impact a microorganism’s ability for growth is its pH. Microbial growth is reduced when pH fluctuates on either side of the optimum range.33 similarly temperature also significantly influence microorganism growth and enzyme yields, particularly in amylase-producing bacteria. In the current study, the highest growth of B. Subtilis was observed at 37 °C and pH 7. Also the growth was increased gradually up to 36 h incubation time and decreased with longer periods. The growth of B. subtilis at different pH ranges, temperature and incubation time was reported by several other studies. The current outcome agrees with the findings of Namo, 10 who observed that the pH of 7, temperature at 37° C and an incubation period of 37 h are the optimum conditions for the growth of Bacillus spp. The highest growth of B. subtilis T9-05 isolated from fish sauce was found to be at pH 7-8 and 35° C.34 The ideal growth conditions for B. subtilis K-C3 isolated from Kapi, were found at pH 8, although it can thrive at pH 6–9 and ideal temperature at 35° C.35
Conclusion
The main goal of this research was to identify and characterize bacteria from soil samples capable of producing amylase enzyme, a crucial protein for industrial processes, thereby filling the gap in the search for efficient amylase producers. Soil samples from Biligiriranga hills in Karnataka were collected and three bacterial isolates out of 15 were identified as amylase producers. The isolates were gram-positive, showed positive for potassium hydroxide, KOH tests and negative for methyl red test. Best amylase producing isolate was subjected to molecular detection and confirmed as Bacillus subtilis, based on 16S rRNA sequencing. The 16S rRNA sequence of B. subtilis strain B has been deposited in NCBI GenBank nucleotide database (Accession No PP112150). B. subtilis (Sample 2) growth was greatly impacted by the pH, temperature and incubation time. The optimum growth conditions for this culture were observed as 37° C, pH 7 and 37h of incubation period. B. subtilis isolated from soil exhibits robust amylase enzyme activity, alongside notable levels of protease and lipase activities. These enzyme activities make it a promising candidate for industrial-scale production of enzymes. Based on the findings of the current investigation, B. subtilis Sample 2 isolated from soil can be used to generate the amylase enzyme on an industrial scale.
Acknowledgement
The authors are grateful to Department of Studies in Biotechnology, University of Mysore for providing the facilities to conduct the experiments.
Funding Sources
The research is self-funded, with all expenses covered personally, ensuring independence and autonomy in project execution.
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Data Availability Statement
The data supporting the findings presented in this manuscript is available upon request from the corresponding author. The 16S rRNA partial sequence has been deposited in the NCBI GenBank as B. subtilis strain B with the accession no. PP112150.
Author’s contribution
Each author participated in drafting and revising the manuscript critically for important intellectual content. Shivalingaiah contributed to the conceptualization, methodology design, data analysis, and oversaw the study and offered guidance throughout the research endeavour. Bindhushree participated in data collection, processing, interpretation, and manuscript drafting, Basavaraju provided expertise in statistical analysis and critical revision of the manuscript, and Pushpalatha participated in conceptualizing the study, designing the methodology, and analyzing the data and supervised the study and provided guidance throughout the research process.
References
- Bhattarai, A., Bhattarai, B., Pandey, S. Variation of soil microbial population in different soil horizons. microbiol. exp. 2015; 2(2): 00044.
CrossRef - Saha, M.L., Nowshin, K., Akter, T., Rahman, I.A., Khan, T. Isolation and identification of amylolytic bacteria from garbage and garden soil. Bangladesh. Bot. 2019; 48(3): 537-545.
CrossRef - Omemu, A.M., Akpan, I., Bankole, M.O., Teniola. Hydrolysis of raw tuber starches by amylase of Aspergillus niger AM07 isolated from the soil. J. Biotechnol. 2005; 4(1): 19-25.
- Morris, S.J., Blackwood, C.B. (2015). Soil Microbiology, Ecology and Biochemistry (4th ed.). (ed. PA Eldor) Academic Press. pp. 273-309.
CrossRef - Gurung, N., Ray, S., Bose, S., Rai, V. A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. Biomed Res.Int. 2013;1-8.
CrossRef - Mohamed, S., Khan, J., Al-Bar, O., El-shishtawy, R. Immobilization of trichoderma harzianum α-amylase on treated wool: optimization and characterization. Molecules. 2014; 19(6): 8027–8038.
CrossRef - Yanhong, L., Jigang, Y., L. Fudong, L., Hui, P., Xuecheng, Z., Yazhong, X., Chao, H. Crystal structure of a raw-starch-degrading bacterial α– amylase belonging to subfamily 37 of the glycoside hydrolase family GH13. Rep. 2017; 7: 44067.
CrossRef - Priyavyar and shirsat, M. Biotechnological aspect of alpha amytnom Bacillus subtilis, Bacillus licheniformis. res. j. med. clin. sci. 2016; 2(7); 1-6.
- Mishra, S., Behera, N. Amylase activity of starch degrading Bacteria isolated from soil receiving kitchen wastes. J. Biotechnol. 2008; 7(18): 3326-3331.
- Namo, F. M. Isolation and characterization of amylase producing Bacteria from soil in Debre Berhan Ethiopia. of Bio. Pharma. and Chem. Res. 2022; 9(3): 48-60.
- Srivathsan, V., Bhandari, M., Swaminathan, P. Isolation and characterization of starch degrading bacteria from disparate soil samples. appl. biol. biotechnol. 2022; 10(5): 193-197.
CrossRef - de Souza, P.M., de Oliveira Magalhaes, P. Application of microbial α-amylase in industry – A review. J. Microbiol. 2010; 41(4): 850-861.
CrossRef - Khassaf, W. H., Niamah, A. K., Al-Manhel, A. J. Study of the Optimal Conditions of Levan Production from a Local Isolate of Bacillus subtilis subtilis w36. Basrah J.Agric.sci. 2019; 32(2): 213-222.
CrossRef - Al-Sahlany, Gddoa, S.T., Al-Asady., Kadhim, A. Poly hydroxy butyrate (PHB) production from local isolates of Bacillus Basrah J.Agric.sci.2015; 28(2): 186-196.
- Al-awsy M., Obiady S.A., Obaidi A.A. Producing of amylase enzyme from thermophilic bacteria using agricultural wastes as a substrate. j. basic appl. Sci. 2017; 11(13): 158-164.
- Parmar, D., Pandya, A. Characterization of amylase producing bacterial isolates. Env. Pharmacol. Life Sci. 2012; 1(6): 42-47.
- Holt, J.G. Bergey’s Manual of Determinative Bacteriology, Lippincott Williams and Wilkins, Baltimore, MA, USA, 1994. 9th edition.
- Bennani, S., Mchiouer, K., Rokni, Y., Meziane, M. Characterization and identification of lactic acid bacteria isolated from Moroccan raw cow’s milk. Greener Biol. Sci. 2017; 25: 4934-4944.
- Abegaz, K. Isolation, characterization, and identification of lactic acid bacteria involved in traditional fermentation of border Ethiopian cereal beverages. J. Biotechnol. 2007; 6 (12): 1469-1478.
- McDevitt, S. Methyl Red and Voges-Proskauer Test Protocols.Washington, DC: American Soc. for Micro. 2009.
- Silaban, S., Marika, D.B., Simorangkir, M. Isolation and Characterization of Amylase-Producing Amylolytic Bacteria from Rice Soil Samples. Journal of Physics: Conf. Series. 2020; 1485(1): 012006.
CrossRef - Nimisha, P., Moksha, S., Gangawane, A.K. Amylase activity of starch degrading bacteria isolated from soil. j. curr. microbiol. appl. sci. 2019; 8(4): 659-671.
CrossRef - Ram Kumar, T., Ramkumar, J., Karthikeyan, S., Ramesh Babu, N.G., Manivasagan, V. (2017). Isolation, optimization, production and purification of alpha amylase from soil bacteria. J. Eng. Technol. 2017; 4(7): 2290-2293.
- Kiran, S., Singh, A., Prabha, C., Kumari, S., Kumari, S. Isolation and Characterization of Thermostable Amylase Producing Bacteria from Hot Springs of Bihar, India. J. Pharma Bio Sci. 2018; 17(2): 28-34.
CrossRef - Vaidya, S., Rathore, P. Isolation, screening and characterization of amylase producing bacteria from soil of potato dump sites from different regions of Madhya Pradesh. Sangavi institute of management and science, Indore, India. Conference: Life Sci. res. j. Int. conf. on rec. tren. in agri. vet. and life sci. 2015; ISBN 978-93-84124-26-7.
- Ghazala, I., Haddar, A., Ben, M., Semia, R. and Chaanouni, E. (2016). Screening and molecular identification of new microbial strains for production of enzymes of biotechnological interest. Braz Arch Biol Technol. 59; 1-12.
CrossRef - Singh, P., and Kumari, P. Isolation and characterization of amylase producing Bacillus from selected soil sample. Int. j. res. biol. sci. 2016; 5(2): 24-29.
- Luang-In, V., Yotchaisarn, M., Saengha, W., Udomwong, P., Deeseenthum, S., Maneewan, K. Isolation and Identification of Amylase-Producing Bacteria from Soil in Nasinuan Community Forest, Maha Sarakham, Thailand. Pharmacol. J. 2019; 12(3): 1061-1068.
CrossRef - Krishma, M., Radhathirumalaiarasu, S. Isolation, identification and optimization of alkaline amylase production from Bacillus cereus using agro-industrial wastes. j. curr. microbiol. appl. sci. 2017; 6(1): 20- 28.
CrossRef - Rakaz, M.A., Hussien, M.O., Ibrahim, H.M. Isolation, Extraction, Purification, and Molecular Characterization for Thermostable α-Amylase from Locally Isolated BacillusSpecies in Sudan. Res. Int. 2021; (2): 1-8.
CrossRef - Tole, S.B., Fawade, M.M., Patil, S.M. Amylase Activity of a Starch Degrading Bacteria Isolated From Soil Around Tulja – Bhavani Temple of Tuljapur. IOSR pharm. biol. sci. 2016; 11(6): 1-6.
- Sangappanavar, A., Nayak, P., Sultana, Q., Pai, A., Venkatesh Kamath, B., Rajesh, K.S. Isolation and characterization of amylase producing bacteria from soil samples. Chem. Bull. 2023; 12(5): 4875-4887.
- Singh, P., Gupta, P., Singh, R., sharma, R. Factors affecting α-amylase production on submerged fermentation by Bacillus Int. j. pharm. life sci. 2012; (3)12: 2243-2246.
- Sanjaya, A.P., Praseptiangga, D., Zaman, M.Z., Umiati, V.F., Baraja, S.I. (2023). Effect of pH, temperature, and salt concentration on the growth of Bacillus subtilis T9-05 isolated from fish sauce. IOP Conf. Ser.: Earth Env. Sci. 2023;1200
CrossRef - Pongsetkul, J., Benjakul, S., Sumpavapol, P., Vongkamjan, K., Osako, K. Bacillus subtilis KC3 isolated from Thai salted shrimp paste (Kapi): Its extracellular enzymes and use as a starter culture in Kapi production. Food Biochem. 2018; 42(6): 3473-3482.
CrossRef

