Introduction
Cultivation plays a vital role in India’s economy and 54.6% of total workforce is connected in cultivation and similar fields1 and accounts for 17.8% of global Gross Value Added (GVA) for year 2019-20. Maximum wastes that are generating from these industries are inedible and produces by many activities such as crop harvesting, animal husbandry and others. According to global annual statistics estimated that the production of rice straw, wheat straw, corn straw, sugarcane bagasse and rice husk generation at 731, 354, 204 181 110 Mt respectively.2 Though wood biomass squander was put at 4.6 Gt/Yr. waste from coffee and olive oil industries were assessed at 7.4 and 30 Mt/Yr. Almost 80% of the total wastes obtained from cereals and sugar cane in total biomass.3, 4, 5 The possible way for the disposal of these wastes are burning in fields or using for landfills in turn which creates many environmental problems. Now a days researchers are exploring these waste materials for production of important compounds in chief and cost effective manner.
The major biological activity of amylase enzyme is to catalyze starch into glucose, maltose and maltodextrines and classified under the family of GH13, with eight subfamilies.6 Apart from this it also engaged in other physiological phenomenon that include germination, defenses and development.7 It is a complex enzyme which includes three enzymes, i.e., α-, β and glucoamylases. Amongst these α-amylase has broad advantages in many industries viz., pharmaceutical, food, textile, paper, and detergent8 (Fig.1) due to its thermal and pH stability.9 Commercial production of microbial enzymes showed approximately 30% of the global market.10 Generally many microorganisms have the potential to secret α-amylase enzyme, among them fungi with maximum potential and appropriate candidate for industrial production. These fungi have the ability to utilize low-cost renewable substrates as a medium for growth, ease of enzyme recovery, pH stability, temperature, and less cofactor.11 Because of low water, less energy utilization and high enzyme yield SSF is more preferable than the submerged fermentation for the commercial production of enzymes.12
In general the most important limiting factor in present usage of commercial enzymes is manufacturing methods is their cost. Growth medium will determine the 30–40% cost of many industrial enzymes.13 Current years, the usage of lignocellulosic biomass in biorefinaries as a renewable waste for the generation of bioenergy, chemicals, and enzymes has gained strength.14 Effective utilization of this waste as cheaper source for the manufacturing of commercially industrial enzymes would not only be useful in decreasing the enzyme cost but also in increasing the value of underutilized substrates. Therefore, increasing the tribal economy with solving dual purpose in mutualistic method. Exploitation of various solid substrates for production of a enzymes with different fermentation methods is well studied.15, 16 However, the hunt for novel substrates for cost–effective production of more robust enzymes is still going on.
 |
Figure 1: Amylase applications in different industries |
Apart from this the other approaches for the enhanced production of important enzymes is use of chemical inducers, the use genetically modified microorganisms, or the co-culture methods are important options. Use of chemical inducers and genetically modified bacteria having its own disadvantages. Co-culturing microbes were further evolved, and found better option for the production of many industrially important products such as pharmaceuticals, nutraceutical, nourishment, and drinks on a huge scale.17, 18 It also plays outstanding role in the bio-remediation and bio-energy divisions.19, 20 Co-culture system was its own advantages over the mono cultures i.e., flexibility, vigor, and ability to attempt modern errands and has immense potential for biotechnological applications.21 Moreover, artificial co-culture methods overcome obstacles of monocultures or consortia with additional focal points in investigating allelopathic relations22 in nourishment industries with fermentation23 and medicate disclosure.24
Table 1: General agro-industrial wastes used as a renewable substrates used in SSF for valorization to value added products.
| S. No | Renewable Substrate | Source Organisms | Product | ||
| Bacterial Co-Culture | Fungal Co-Culture | Both Bacteria And Fungi Co-Culture | |||
| 1. | Apple pomace | Aspergillus ornatus, Alterneria alternata | Citric acid | ||
| 2. | Sweet potato flour | Trichoderma sp and Sacharomyces cerevisiae | Bioethanol | ||
| 3. | Paddy straw | Endoglucanase, Xylanase Filterpaparase and β-glucosidase | |||
| 4. | Wheat bran | Bacillus thuringiensis and Bacillus cereus | Xylanase | ||
| 5. | Palm kernel cake | Bacillus-Tricoderma | cellulase | ||
| 6. | Corn cob and Bermuda grass | Tricoderma reesei and Aspergillus niger GS1 | Amylase, Fpase and Xylanase | ||
| 7. | Saw dust and Wheat bran | Cladosporium Sphaerospermum, Aspergillus flavus and Epicoccum purpurascens | Xylanase | ||
| 8. | Wheat bran | Aspergillus penicillioides and Aspergillus flavus | Xylanase, Fpase, β-xylosidase CMCase. | ||
| 9. | Wheat bran pulse husk and mustered peel | Trametes hirsute and Phaenerochaete sp. | Laccase, Pectinase | ||
| 10. | Pineapple peel banana peel, and papaya peels | Phaenerochaete chrysosporium and Scizophyllum communne | α-Amylase and Cellulase | ||
The substrates listed in table 1 are easily available and also a nutrients rich agro industrial waste materials which can be used as substrates in SSF. The choice of considering pretreated substrates for enzyme production has an impact on the outcome. Many fungal strains have the ability to produce α-amylase. The glucoamylase was produced by different organisms like Aspergillus awamori, A. saitoi, A. oryzae, Rhizopus sp, Mucor sp, Penicillium sp., and Yeast.25 Hence, the present study was intended to choose natural waste as cheaper source for the production of robust and low cost amylase enzyme suitable for industrial applications. Further, production capacity of amylase enzyme among the two indigenous Aspergillus strains individually and in co-culture methods using the SSF and SmF of natural agro-industrial waste as carbon source.
Materials and Methods
Microorganism
An indigenous two Aspergillus strains (Aspergillus unguis Accession number KX816008 and Aspergillus protuberus Accession number KX427028) were evaluated for their capability to utilize solid waste as a carbon source for its multiplication and production of amylase under SSF conditions. Czapek Dox agar medium was used to maintain pure cultures at 30 ± 2°C and maintain at 4°C.
Inoculum preparation
10-15 ml of sterile distilled water with 0.8% tween-80 was added to the slant which was well speculated and shaken vigorously, then the spore count was adjusted to 1x 107 spores/ml with fresh sterile distilled water and used to carry out solid state fermentation of different substrates.
Plate Screening Method
The selected fungal strains were spot inoculated on the medium which is supplemented with starch and incubated at
30 ± 2°C . Dominant growth was observed for both the cultures at 48h of incubation and 1% iodine solution was poured on to the culture plate and incubated (Karri et al. 2014). Vanishing of blue color surrounding the colony indicated the production of extracellular amylase enzyme which break down the starch in to glucose and utilized for the physiological activities. In the culture medium where the starch is not degraded showed in the blue color only.
Lignocellulosic substrates
Commonly available local agro-industrial waste substrates were collected, dried under controlled temperature and ground in to fine powder individually and sieved with a 2 mm screen to get even particle size. In the present study we included rice husk, groundnut fodder, saw dust, sugarcane bagasse and castor husk were chosen.
Solid State Fermentation
250 ml Erlenmeyer flasks was used to carry out Solid state fermentation (SSF) which contains 10 grams of different lignocellulosic substrates in each flask. Czapek Dox broth with the composition of two gm of NaNO3, one gm of K2HPO4, 0.5 gm of MgSO4 7H2O, 0.5 gm of KCl, 0.01 gm of FeSO4 7H2O, 30 gm of sucrose and 5 gm of starch. 50% moisturization was maintained in each carbon source by adding different volumes of (10-15ml) sterile dH2O and sterilized at 121˚C for 15 min. Medium was inoculated with specified volume of fungal spores and incubated at ambient temperature and samples were collected at every 24 h for enzyme estimation by slightly modified Miller28 method .
Co-culture method by Submerged Fermentation
In co-culture system SmF was done in 250 ml conical flasks. Ten grams of rice husk was used as a substrate and maintained 50% moisture level by adding 10 ml of distilled water and then sealed with cotton plug and autoclaved. 7-day-old mycelia were used for the preparation of the inoculum. Different spore ratios (1:1, 1:3, 3:1 and 0.5:0.5) were inoculated and incubated at ambient temperature (30 ± 2oC) 100 rpm/ min for 10 days. The samples were collected at every 24 hours for processing.
Enzyme extraction
Amylase enzyme was recovered from the fermented substrate by adding different volume of sterile distilled water and shaking for 30 min in shaking incubator at a solid to substrate ratio of 1:10, then the extract was filtered with whatman no 1 filter paper and centrifuged at 5000 rpm for 20 min to obtain clear filtrate26, 27 and used as a crude enzyme to measure its activity by spectrophotometric method.
Enzyme assay
One International Unit is expressed as one μmol of glucose equivalents released per minute per ml. Appropriate dilutions were used in assessment of enzyme activity. Alpha and glucoamylase activity was estimated by the protocol of Miller.28
α- amylase activity
1% soluble starch was used as a substrate in 0.1M citrate buffer pH 5.0. Freshly prepared culture filtrate was used as a enzyme source. The alpha amylase activity was measured by adding 1ml of diluted enzyme solution and 1ml of buffer solution along with the appropriate amount of substrate in a clean glass tube and incubated for 30 min at 45oC. After incubation period 3 ml of 3, 5- dinitrosalicylic acid (3,5-DNS) reagent was added to the tube to stop the reaction and tubes were boiled for 15 min in boiling water bath. Before cooling to room temperature, 1ml of 40% Rochelle salt was added to each tube to preserve its color. Final volume in all tubes was made upto 7 ml with distilled water. Absorbance was measured at 575 nm with UV-Visible spectrophotometer considering 0.1 M citrate buffer as a reference blank and compared results with standard curve.
Glucoamylase activity
1% maltose in 0.1 M citrate buffer (pH 5.0) was used as a substrate for the estimation of glucoamylase activity at 450C for 30 min. The enzymatic reaction was carried out by pipetting 1 ml of diluted enzyme and one ml of citrate buffer in appropriate amount of substrate and incubated at 450C for 30 min. 3 ml of 3, 5-DNS reagent was added to the test tube to stop enzymatic reaction and the contents of the tube were heated for 15 min in boiling water bath. Activity was estimated as mentioned in alpha amylase activity.
Protein determination
Lowry method29 was employed for the estimation of ES proteins secreted by two strains into the culture medium after specific incubation periods.
Statistical analysis
Triplicates were analyzed by using Descriptive Statistics in MS-EXCEL-2007 software.
Results and Discussion
A. protuberus and A.unguis were indigenous fungal cultures that was isolated from the Mahanandi forest soil sample and soil samples procured from the Kadapa cotton ginning mill respectively and both of these cultures were well known for hyper cellulase enzyme production. In this article compared these cultures for their ability to produce another industrial important enzyme i.e., amylase in co –culture system and also screened the agro industrial waste for the selection of suitable substrate for low cost production of the amylase enzyme. The results of screening of fungal cultures are shown amylase production on plates. Maximum zone of starch hydrolysis was seen on the plates of Aspergillus protuberus and Aspergillus unguis indicated that are amylase producers (Fig.2a, b).
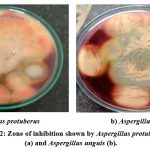 |
Figure 2: Zone of inhibition shown by Aspergillus protuberus (a) and Aspergillus unguis (b). |
Screening of agro-industrial wastes
Globally (urban and rural locations), clearance of solid waste is a troublesome and persistent problem. The general way of disposal of this solid waste is incineration or landfill dumping. In recent years several scientists were studied on generation of important products from this waste in ordered to reduce the natural issues.30 Several value-added products were generated from the glycerol waste which is produced from biodiesel industry, i.e., Organic acids (citric acid, succinic acid), and other intermediate products (ethanol, and intermediate additives), in order to produce biopolymers by means of microbes.31, 32, 33 Exploitation of biological techniques in waste management is becoming a important method in feasible improvement.32, 34
Solid state fermentation (SSF) is an elective approach, for converting agro-industrial squanders and its byproducts into value-added items, i.e., bioactive substances, bioplastics, and biofuels,35 because it mimics the natural environment. Generally, SSF utilizes waste generated from the agriculture as a medium of low or no financial esteem; instead, their transfer is natural concern. Raw material availability and its cost are the two significant limitations need to be well thought-out while selecting a raw material in SSF.36 The selected raw material should support the utmost growth of microorganism and as well as high product yield. In the present study, various agro-industrial wastes including rice husk, castor husk, sawdust, sugarcane bagasse and groundnut husk were used for the solid state fermentation. All the five were found to be good substrates as the α- amylase, glucoamylase and protein content activity was observed.
Maximum yield of α-amylase on all solid substrates except castor husk and groundnut waste was registered on second day of incubation. Higher yield of α-amylase (1.614 U/g of substrate & 0.958 U/g of substrate) was showed on 2nd day on rice husk by A.protuberus and A.unguis, whereas less yield of α-amylase (0.511 U/g of substrate) was noted on 2nd day when sawdust used as a substrate by A.protuberus. Respectively groundnut husk was showed less yield of α-amylase activity (0.580 U/g of substrate) on 5th day. All solid substrates were showed maximum α-amylase activity on 2nd day of incubation except groundnut fodder by A. unguis (Fig.3 &5).
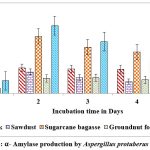 |
Figure 3: α- Amylase production by Aspergillus protuberus in SSF. |
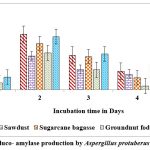 |
Figure 4: Gluco- amylase production by Aspergillus protuberus in SSF. |
Maximum titres of glucoamylase on all solid substrates like sugarcane bagasse, rice husk, castor husk and sawdust was showed on the second day of incubation with A. protuberus. Among solid substrates tested, castor husk yielded highest activity of 0.692 U/g of solid substrate as against 0.665 U/g of solid substrate by rice husk on 2nd day of incubation. Growth of A. protuberus lowest activity of glucoamylase on groundnut husk on 5th day of incubation. By using A.unguis on production of glucoamylase was high in sugarcane bagasse, rice husk and groundnut fodder on 1st day of incubation, similarly remaining two substrates showed on 5th day of incubation. Groundnut waste was observed highest production of glucoamylase (0.847 U/g of substrate) on 1st day followed by rice husk yield of 0.828 U/g of substrate and very less activity of glucoamylase (0.455 U/g of substrate) on 5th day (Fig.4 & 6).
Among the solid substrates used in the present study, maximum secretion of extracellular protein (2.259 and 1.898 mg/g of substrate) on castor husk at peak time interval on 2nd day and 5th day of incubation was recorded in A.protuberus and A.unguis. Whereas the protein content was low (0.253 and 0.245 mg/g of solid substrate and) on rice husk at peak time interval on 3rd & 1st day of incubation by A.protuberus and A.unguis (Fig.7 & 8).
Bindu naik et.al.37 screened various agro-waste for the production of pullulanase enzyme from the endophytic Aspergillus sp. and found that wheat bran was found to be the best substrate with maximum yields of 65.33± 2.08U/gds. Shruthi et.al.,38 screened locally available many agro industrial waste material and observed the considerable production of FPase (5.9 FPU/g of substrate), CMCase (1.1 U/g of substrate) and β-glucosidase activity (6.5 U/g of substrate) and extracellular protein (27.0 mg/g of substrate) in SSF using ground nut fodder. Three different solid wastes viz., sugarcane bagasse, sawdust, and tea residue was screened in SSF and demonstrated that sugarcane baggage was the excellent solid support among the diverse solid substrates used in this study for higher generation of cellulase and amylase. Sugarcane bagasse backed surprising generation of FPase (15.42 FPU/g of substrate), CMCase (17.89 U/g of substrate) and β-glucosidase (2.43 U/g of substrate), and amylase activity (15.12 U/g of substrate) in SSF. Noteworthy secretion of protein (45.75 mg/g of substrate) on sugarcane bagasse was taken note.39 In spite of the fact that, production of ethanol from lignocellulosic substrates in a cost effective way, it is a vital challenge with the commercialization of the production practice.
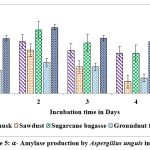 |
Figure 5:. α-Amylase production by Aspergillus unguis in SSF. |
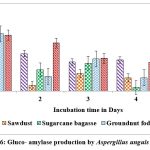 |
Figure 6: Gluco- amylase production by Aspergillus unguis in SSF. |
Co-culture method
Co-cultured microorganisms have advantageous over the individual cultures as the synergistic action of metabolic way of organisms which naturally upgrade the production of commercial enzymes. In nature organisms are not survived as pure cultures and also microbes belong to identical genus encompass the better compatibility with each other, hence, widely used to produce different metabolites. This can be concluded from reports on pair of microbes such as Bacillus cereus and B. thuringiensis, Aspergillus niger MS23, and A. terreus MS105, Clostridium thermocellum ATCC 27405 and C.beijerinckii ATCC 51743, Aspergillus flavus and A. penicillioides.40, 41, 42, 43 Maximum levels of α-amylase ( 12 u/g of substrate) was record on 2nd day of fermentation when inoculated both cultures at 1:1 ratio and the enzyme secretions are gradually decrease when the incubation period is increasing from 2nd day to 10th day (Fig. 9). Dreaded amount of α-amylase was observed when inoculated these cultures at 1:3 ratio on 10th day of incubation. Maximum levels of gluco-amylase (3 u/g of substrate) was observed on 2nd day of fermentation when inoculated both cultures at 0.5: 0.5 and the enzyme secretions are gradually decrease when the incubation period is increasing from 2nd day to 10th day (Fig. 10). Dreaded amount of gluco-amylase was observed when we inoculated these cultures at 0.5:0.5 ratio at 10th day of incubation.
Among the combinations used in the present study, 1:3 combination was good for the production of highest concentration of extra cellular protein (20 mg/g of substrate) on rice husk on 2nd day of incubation. Whereas the 0.5:0.5 combination was the poor combination to produce extracellular protein content (2.5 mg/g of solid substrate) on rice husk at peak time interval on 10th day of incubation (Fig.11).
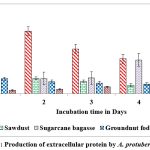 |
Figure 7: Production of extracellular protein by A. protuberus in SSF. |
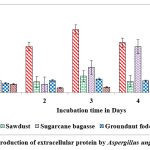 |
Figure 8: Production of extracellular protein by Aspergillus unguis in SSF. |
In co-culture study Ankit lodha et.al.,44 observed the elevated levels of cellulase enzyme (6.71 FPU/gds) when steam pretreated wheat bran was supplemented with 106 spores of each of the Trichoderma reesei and Penicillium citrinum in SSF at 30 °C , pH 5 and 70% moisture content on 6th day of incubation. In an another co-culture study Saccharomyces cerevisiae Y3401 and Wickerhamomyces anomalus Y3604 was employed for the utmost production of ethyl acetate i.e., 6.4 g/l at the ratio of 3:1 than in individual fermentation.45 Ledys et.al.,46 used triple co–culture study for the enhanced production of ligninolytic enzymes and to decolorize Reactive black dye and found that the inoculation of 1000 μL of each culture of T. viride and A. terreus into the 7–day culture of Leptosphaerulina sp is the best combination than monoculture and previously used chemical inducers.
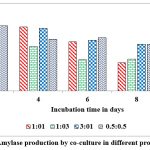 |
Figure 9: α-Amylase production by co-culture in different proportions in SSF |
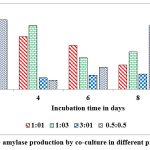 |
Figure 10: Gluco- amylase production by co-culture in different proportions in SSF. |
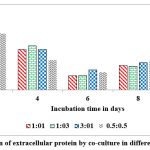 |
Figure 11: Production of extracellular protein by co-culture in different proportions in SSF. |
Conclusion
Production of low cost amylase enzyme is the main goal of the present study. Hence various agricultural residues were studied to carry out SSF by Aspergillus sp and demonstrated that the studied Aspergillus strains has the potential to utilize agricultural waste residues for the production of α-amylase, glucoamylase enzyme and protein content individually. The enzyme productions and extra cellular protein was drastically increased in co-culture study. Hence, concluded that co-culture method is the best method for the enhanced production of amylase enzyme in SmF on rice husk by Aspergillus strains in low cost manner.
Acknowledgment
We would like to express our sincere gratitude to all the authors, they have contributed to the publication of this research paper. This research has not received any funding from any organization.
Conflict of Interest
The authors have no financial or personal conflict of interest.
References
- Census 2011 https://agcensus.nic.in/document/ac/air2010-11%20complete_compressed.pdf
- Cho E.J., Trinh L.T., Song Y., Lee Y.G., Bae H.J. Bioconversion of biomass waste into high value chemicals. Bioresource Technology. 2020; 298:122386.
CrossRef - IEA (International Energy Agency). Mobilization of agricultural residues for bioenergy and higher value bio-products: Resources, barriers and sustainability. IEA Bioenergy Task. 2017; 43:1.
- Lal R. World crop residues production and implications of its use as a biofuel. Environment International. 2005; 31(4):575-584.
CrossRef - UCS (Union of Concerned Scientists). Turning Agricultural Residues and Manure into Bioenergy. Cambridge: UCS; 2014.
- Liu Y., Lei Y., Zhang X., Gao Y., Xiao Y., Peng H. Identification and phylogenetic characterization of a new subfamily of α-amylase enzymes from marine microorganisms. Marine Biotechnology. 2012;14:253-60.
CrossRef - Sugimura Y., Michiyama H., Hirano T. Involvement of α-Amylase Genes in Starch Degradation in Rice Leaf Sheaths at the Post-Heading Stage. Plant Production Science. 2015; 18:277–283.
CrossRef - Jujjavarapu S.E., Dhagat S. Evolutionary trends in industrial production of alpha-amylase. Recent Patents on Biotechnol. 2019; 13(1):4–18.
CrossRef - Balakrishnan M., Jeevarathinam G., Kumar S.K.S., Muniraj I., Uthandi S. Optimization and scale-up of α-amylase production by Aspergillus oryzae using solid-state fermentation of edible oil cakes. BMC Biotechnology. 2021, 21, 33.
CrossRef - Rajagopalan G., Krishnan C. Alpha-amylase production from catabolite derepressed Bacillus subtilisKCC103 utilizing sugarcane bagasse hydrolysate. Bioresource Technology. 2008; 99:3044–3050.
CrossRef - Sethi B.K., Jana A., Nanda P.K., DasMohapatra P.K., Sahoo S.L, Patra J.K. Production of alpha-Amylase by Aspergillus terreusNCFT 4269.10 Using Pearl Millet and Its Structural Characterization. Frontiers in Plant Science. 2016; 7:639.
CrossRef - Soccol C.R., Da Costa E.S., Letti L.A., Karp S.G., Woiciechowski A.L, de Souza Vandenberghe L.P. Recent developments and innovations in solid state fermentation. Biotechnology Research and Innovation. 2017;1(1):52-71.
CrossRef - Joo H, Chang C. Production of protease from a new alkalophilic Bacillus I.312 grown on soya-bean meal: optimization and some properties. Process Biochemistry. 2005;40:1263–1270.
CrossRef - Banu J.R., Kavitha S., Tyagi V.K., Gunasekaran M., Karthikeyan O.P., Kumar G. Lignocellulosic biomass based biorefinery: A successful platform towards circular bioeconomy. Fuel. 2021:15;302:121086.
CrossRef - Gupta R., Gigras P., Mohapatra H., Goswami V.K., Chauhan B. Microbial -amylase: a biotechnological perspective. Process Biochemistry. 2003: 38, 1599–1616.
CrossRef - Sivaramakrishnan S., Gangadharan D., Nampoothiri K.M., Soccol C.R., Pandey A. Amylases from microbial sources – an overview on recent developments. Food Technology and Biotechnology. 2006:44, 173–184.
- Bader J., Mast‐Gerlach E., Popović M.K., Bajpai R., Stahl U. Relevance of microbial coculture fermentations in biotechnology. Journal of Applied Microbiology. 2010:09(2):371-87.
CrossRef - Ma Q., Zhou J., Zhang W., Meng X., Sun J., Yuan Y.J. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PloS One. 2011:7;6(10):e26108.
CrossRef - Breugelmans P., Barken K.B., Tolker-Nielsen T.et al. Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiology Ecology. 2008;64(2):271–282
CrossRef - Angenent L.T., Karim K., Al-Dahhan M.H., et al.Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends in Biotechnology. 2004; 22(9):477–485.
CrossRef - Hays S.G., Patrick W.G., Ziesack M., Oxman N., Silver P.A. Better together: engineering and application of microbial symbioses. Current Opinion in Biotechnology. 2015: 1;36:40-9.
CrossRef - DellaGreca M., Zarrelli A., Fergola P., Cerasuolo M., Pollio A., Pinto G.Fatty acids released by Chlorella vulgaris and their role in interference with Pseudokirchneriella subcapitata: experiments and modelling. Journal of Chemical Ecology. 2010; 36:339-49.
CrossRef - Amaral Santos C.A.d., da Silva Libeck B., Schwan R.F.Co-culture fermentation of peanut-soy milk for the development of a novel functional beverage. International Journal of Food Microbiology. 2014; 186:32–41.
CrossRef - Bertrand S., Azzollini A., Schumpp O., Bohni N., Schrenzel J., Monod M., Gindro K., Wolfender J.L. Multi-well fungal co-culture for de novo metabolite-induction in time-series studies based on untargeted metabolomics. Molecular BioSystems. 2014; 10(9):2289-98.
CrossRef - Pandey A., Webb C., Soccol C.R., Larroche, C. Enzyme Technology. Asiatech publishers, New Delhi, Inc.2005: 197.
CrossRef - Deswal D., Khasa Y.P., Kuhad R.C. Optimization of cellulase production by a brown rot fungus Fomitopsis RCK2010 under solid state fermentation. Bioresource Technology. 2011;102(10):6065-72.
CrossRef - Chandra M.S., Viswanath B., Reddy B.R. Cellulolytic enzymes on lignocellulosic substrates in solid state fermentation by Aspergillus niger. Indian Journal of Microbiology. 2007;47:323-8.
CrossRef - Miller G. L. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Analytical Chemistry. 1959:31, 426–428.
CrossRef - Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with Folin Phenol reagent. Journal of Biological Chemistry, 1951:193: 265-275
CrossRef - Chilakamarry C.R., Sakinah AM.., Zularisam A.W., Pandey A., Vo D.V. Technological perspectives for utilisation of waste glycerol for the production of biofuels: A review. Environmental Technology & Innovation. 2021;24:101902.
CrossRef - Chilakamarry C.R., Mahmood S., Saffe S.N., Arifin M.A., Gupta A., Sikkandar M.Y., Begum S.S., Narasaiah B. Extraction and application of keratin from natural resources: a review. 3 Biotech. 2021:11:1-2.
CrossRef - Sirohi R., Pandey J.P., Gaur V.K., Gnansounou E., Sindhu R. Critical overview of biomass feedstocks as sustainable substrates for the production of polyhydroxybutyrate (PHB). Bioresource Technology. 20201; 311:123536.
CrossRef - Sirohi R., Lee J.S., Yu B.S., Roh H., Sim S.J. Sustainable production of polyhydroxybutyrate from autotrophs using CO2 as feedstock: challenges and opportunities. Bioresource Technology. 2021:341:125751.
CrossRef - Liu H., Qin S., Sirohi R., Ahluwalia V., Zhou Y., Sindhu R., Binod P., Singhnia R.R., Patel A.K., Juneja A., Kumar D. Sustainable blueberry waste recycling towards biorefinery strategy and circular bioeconomy: A review. Bioresource Technology. 2021;332:125181.
CrossRef - Sharma P., Gaur V.K., Sirohi R., Varjani S., Kim S.H., Wong J.W. Sustainable processing of food waste for production of bio-based products for circular bioeconomy. Bioresource Technology. 2021;325:124684.
CrossRef - Chimata N.K., Sasidhar P., Challa S. Production of extracellular amylase from agricultural residues by a newly isolated Aspergillus species in solid state fermentation. African Journal of Biotechnology. 2010;9(32):5162-9.
- Naik B., Goyal S.K., Tripathi A.D., Kumar V. Screening of agro-industrial waste and physical factors for the optimum production of pullulanase in solid-state fermentation from endophytic Aspergillus Biocatalysis and Agricultural Biotechnology. 2019;22:101423.
CrossRef - Shruthi K., Yadav P.S., Prasad B.S., Chandra M.S. Cellulase production by Aspergillus unguis in solid state fermentation. Journal of Forestry Research. 2019;30:205-12.
CrossRef - Chandra M.S., Lokesh T., Anusha C., Yadav P.S. Indian Journal of Advances in Chemical Science. Indian Journal of Advances in Chemical Science. 2019;7(3):70-5.
- Abdullah R., Naeem N., Aftab M., Kaleem A., Iqtedar M., Iftikhar T., Naz S. Enhanced production of alpha amylase by exploiting novel bacterial co-culture technique employing solid state fermentation. Iranian Journal of Science and Technology, Transactions A: Science. 2018;42:305-12.
CrossRef - Rehman S., Aslam H., Ahmad A., Khan S.A., Sohail M. Production of plant cell wall degrading enzymes by monoculture and co-culture of Aspergillus niger and Aspergillus terreus under SSF of banana peels. Brazilian Journal of Microbiology. 2014: 45(4), 1485–1492.
CrossRef - Yao W., Nokes S.E. First proof of concept of sustainable metabolite production from high solids fermentation of lignocellulosic biomass using a bacterial co-culture and cycling flush system. Bioresource Technology. 2014:173, 216–223.
CrossRef - Zhao B., Al Rasheed H., Ali I., Hu S. Efficient enzymatic saccharification of alkaline and ionic liquid-pretreated bamboo by highly active extremozymes produced by the co-culture of two halophilic fungi. Bioresource Technology. 2021:319, 124115.
CrossRef - Lodha A., Pawar S., Rathod V. Optimized cellulase production from fungal co-culture of Trichoderma reesei NCIM 1186 and Penicillium citrinum NCIM 768 under solid state fermentation. Journal of Environmental Chemical Engineering. 2020;8(5):103958.
CrossRef - Fan G., Teng C., Xu D., Fu Z., Minhazul K.A., Wu Q., Liu P., Yang R., Li X. Enhanced production of ethyl acetate using co-culture of Wickerhamomyces anomalus and Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering. 2019;128(5):564-70.
CrossRef - Copete-Pertuz L.S., Alandete-Novoa F., Placido J., Correa-Londono G.A., Mora-Martínez A.L. Enhancement of ligninolytic enzymes production and decolourising activity in Leptosphaerulina by co–cultivation with Trichoderma viride and Aspergillus terreus. Science of the Total Environment. 2019;646:1536-45.
CrossRef

