Introduction
Bananas and plantains are a perennial crop, large, herbaceous, monocotyledonous1 that contain an underground stem, and belong to the Musa genus of the Musaceae family. This family of more than 50 species, originate predominantly in Southeast Asia, specifically in the Indo-Malesian, Asian, African, and Australian tropics,2 being known in the Medi-terranean after the conquest of the Arabs in the year 650 AD. The species arrived in the Canary Islands in the 15th century and was brought to America in 1516.3 Among the latter, bananas and plantains are important crops in tropical and subtropical countries4 and represent the fourth most important food worldwide after rice, wheat, and corn.5 The banana contains a low percentage of proteins (1.2%) and lipids (0.3, highlighting its high content of carbohydrates (20%). In the immature banana, the primary carbohydrate is starch.
Starch is converted to sucrose, glucose and fructose when the fruit reaches ripening. It also contains inulin and other compounds called fructooligosaccharides that are not digestible by the enzymes present in the intestine, which also reach the final part of the intestine and have beneficial effects on intestinal transit itself. Additionally, it is a source of potassium that contributes to muscle function and is also a source of vitamin B6.6
The banana is considered the first globalized product in the world and is the fruit with the largest export market, in addition to being the most consumed.7 It is estimated that there are more than 1,000 varieties of bananas worldwide, which provide essential nutrients to the population. The most commercialized variety is the Cavendish, which represents a little less than half of world production, with an estimated 50 million tons. Bananas are very important in underdeveloped, low-income countries with a lack of food, where they help maintain food security as a basic food, and contribute to the generation of economic income as a crop of high commercial value. The global banana trade has developed at unprecedented levels in recent years, with an estimated export volume of 21 million tons in 2019. Key trade drivers include strong supply growth in Ecuador and the Philippines as main exporting countries, and a significant increase in import demand, particularly from China and the European Union.8 The global production of bananas for 2021 was 124,978,578.48 tons in a harvested area of 5,336,862.00 hectares. In Puerto Rico, it was estimated that this year’s production was 77,470.60 tons in a harvested area of 1,559.0 hectares.9 There are many economically important varieties of Musa spp. in Puerto Rico despite not being originally from the Americas. Both the dietary and the economic importance of these crops on the island have led to studies focused on the preservation of varieties and the development of new ones. Furthermore, Puerto Rico acts as an important reservoir of Musa spp., mainly through the efforts of the USDA-ARS Tropical Agricultural Research Station (TARS) and the University of Puerto Rico (UPR) Agricultural Experimental Station. By 2016, the USDA-ARS TARS developed the “Catalog of Musa Accessions Maintained at the USDA-ARS Tropical Agriculture Research Station.” This catalog has over 130 plant accessions held by USDA-ARS TARS. These accessions consist of individuals with origins from Malaysia, Papua New Guinea, Borneo, Taiwan, Eastern Africa, Congo, Cameroon, Honduras, Brazil, and Nigeria, among many others, whose origins are unknown.10
Musa spp. crops can be susceptible to a wide variety of viral, bacterial, and fungal etiology diseases. In this sense, one of the most worrying diseases worldwide today is the Fusarium Tropical Race 4 (TR4) fungus, a disease that affects banana plants and is currently present in 21 banana-producing countries. According to the cases reported, once a cropland has been contaminated with TR4, disease management is difficult and costly.
The importance of the different banana diseases will be discussed with particular emphasis on Fusarium oxysporum f.sp. cubense tropical race 4 (Foc TR4), its implications, and the latent risk it represents for the producing regions of this crop, as well as new tools to predict its presence in risk areas such as the Puerto Rican island.
Genetic variability in Musa spp
Genetic and morphological characteristics
Musa species consist of 11 chromosomes.11 Edible crops are a union of the wild subspecies Musa acuminata and Musa balbisiana which are referred to as genomes A and B, respectively.12 Although hybridization has produced diploid (AA, AB, BB) and tetraploid (AAAA, AAAB, AABB, ABBB) varieties,13 the varieties grown commercially are triploid (AAA, AAB, ABB),14 Some cultures such as the AAA banana varieties can be eaten raw when ripe, while AAB (plantain) or ABB (banana) need to be cooked before being ingested.2
Musa grown in Puerto Rico
Once the agricultural importance of the plantain and banana industry for Puerto Rico is established, it is not surprising that there are a large number of economically important varieties of Musa spp, in Puerto Rico despite not being originally from the Americas, but from Southeast Asia and the Indian subcontinent.15 The island, consequently, thanks to the studies carried out in Puerto Rico in the search for the preservation of varieties and the development of new ones, Puerto Rico is an important reservoir of Musa spp, mainly through the efforts of the USDA-ARS Tropical Agricultural Research Station (TARS) and the University of Puerto Rico (UPR) Agricultural Experimental Station10 sampled Musa spp. plants. In this sense, in Puerto Rico we can find Musa spp. with diverse genomic composition. García16 characterized Fusarium species in Puerto Rico (Isabela, Mayagüez, Gurabo and Aguada). Their sampled Musa spp. had A.B., AAA, AAB, ABB, and AABB genomes; 5 of the 8 genomic compositions of agricultural importance. However, collections present in Puerto Rico show additional genomic compositions (A.A., B.B., AAAB).17,18,19
By 2009 the USDA-ARS TARS repository contained some 135 accessions; however, the genomic composition was unknown for most of them. Thus, a study carried out by Irish and collaborators in 2009 elucidated the individuals in the collection at the level of polyploidy and genomic composition, determining the great variety that is possessed, for more information you can review the work of Irish18 where it refers to the USDA-ARS TARS collection, however, other research has been carried out on the island where variants not mentioned in this work have been imported and used, such as FHIA-01 (AAAB) and FHIA-25 (AAB).19
The varieties of Musa spp. present both in collections and research has already been established. Additionally, USDA-ARS TARS developed the “Catalog of Musa Accessions,” where the 148 available plants in 2016 can be found. The collection has individuals with origins from Malaysia, Papua New Guinea, Borneo, Taiwan, East Africa, Congo, Cameroon, Honduras, Brazil, Nigeria, among many others whose origins are unknown. In addition, it is worth mentioning that there are varieties developed in Puerto Rico such as TARS 17164 – ‘1-A’, TARS 17125 – ‘2-A’, TARS 17154 – ‘3-A’, TARS 17135 – ‘5-A’ TARS 17130 – ‘6-A’, TARS 17159 – ‘8-A’, TARS 17131 – ‘2-R-2, 500’, TARS 17141 – ‘3-R-2, 500’, TARS 17151 – ‘4-R-2 , 500 ‘, TARS 17152 – ’10 -A’, TARS 17179 – ‘Corozal Selection 25’ (se-lections made by the University of Puerto Rico at the Corozal Experimental Station) and TARS 18061 – ‘Johnson’.10 In summary, Puerto Rico is an important reservoir of different species and varieties of Musa spp. Therefore, their management is imperative, specifically referring to the established policies to control or prevent the arrival of potential pests to the Island. In addition, due to this important genetic variability in Puerto Rico, this working group is working on projects related to the development of new resistant varieties from which we hope to have results in the short term.
Musa spp diseases
The Musa agricultural industry and scientists have looked into species that have developed mechanisms that protect them from abiotic and biotic factors. For example, M. balbisiana has developed resistance to pests, drought, and diseases within its genotype. Similarly, M. acuminata produces phytochemical compounds capable of treating diseases.2 However, despite these mechanisms, Musa spp. crops can be negatively affected by many diseases of biological origin, such as viruses, bacteria, and/or fungi.20
Viral disease
For example, the Banana Streak Virus (BSV), Cucumber Mosaic Virus (CMV), Banana Bract Mosaic Virus (BBMV), and Banana Bunchy Top Virus (BBTV) 21 are viruses of greatest concern. Moreover, there are viruses of lower infectivity in these plants, such as Abaca Bunchy top, Abaca Mosaic, Banana Mosaic, Banana Mild Mosaic, and Banana Virus X.22 Among the aforementioned viruses, this review will address those that have the greatest negative impact on Musa sp. The BBTV virus causes the most significant economic losses in the old world, where losses in yields have reached up to 100%.23 The presence of this virus has not been identified in the Americas, although it has been identified in Hawaii,24 and all species of the family of this plant are susceptible to it. In the case of BBMV, it has caused significant economic losses in India and the Philippines, and it has also been reported in Colombia and Costa Rica, indicating that it is already near the Caribbean islands. Furthermore, the virus is transmitted by aphids. In the Puerto Rico area, the presence of BSV has been identified in the same way in other nearby countries such as Brazil, Colombia, Costa Rica, Ecuador, Cuba, Haiti, Honduras, Jamaica, Nicaragua, Venezuela, the state of Florida (USA) and the Dominican Republic.20,22, 25 It is worth mentioning that this virus has caused losses of up to 90% in the Ivory Coast area. However, in many affected areas, its damage appears to be slight.20 Analyzing the recent literature regarding the identification of viruses in Musa sp. since 2016, we can find new reports such as BBTV in Togo,26 and in South Africa;27 CMV in Musa x paradisiaca cv Chini Champa in Northeast India28 and Banana mild Mosaic virus in India.29 Solving problems due to viruses causes an increase in production costs. For example, an investigation by Selvarajan et al. (2017) determined that applying 125 to 150% more fertilizers than average was essential to maintain the yields of the French plantain cv. Nendran that were infected with BBMV.30
Bacterial disease
One of the diseases of bacterial origin found in Musa spp is the moko. The bacterium Ralstonia solanacearum, the causal agent of this disease, triggers pseudostem tissue rotting. Its dissemination can occur through plant propagation, aphids, machinery, water in irrigation systems, and the movement of people.31 Although this bacterium has not been recorded affecting crops in Puerto Rico, it has been identified in countries of America such as Honduras, Panama, Costa Rica, Belize, Colombia, Ecuador, El Salvador, Granada, Nicaragua, Suriname, Venezuela, Brazil, Peru, Guatemala, Jamaica, and part of Mexico.20 Another disease of bacterial origin is the bacterial wilt of plantain, which is caused by Xanthomonas campestris pv. musacearum. This disease, identified in East Africa, has spread in this continent due to its easy dispersal by insects that visit the flowers. The bacterial wilt of plantain is also easily transmitted by contact with work tools. Some areas have experienced a total loss of production because of the fruit rotting that this bacterium causes.20,32 Furthermore, the bacteria Erwinia spp. causes a disease characterized by rotting in the pseudostem, generating an exudate with a bad smell, and bending of the stem.33 One of the Erwinia species identified in Puerto Rico is E. chrysanthemi, which is the cause of the soft rot that causes abortion of the bunch. This bacterium was isolated from the banana of the Hua Moa variety.34 Finally, recent research determined the presence of Klebsiella variicola in Haiti. This bacterium causes soft rot in bananas and other plants, such as carrots. Therefore, there could be a large number of bacteria that can cause damage to Musa spp. plants and especially close to the Puerto Rico area.35
Phytopathogenic fungi
The main concern for the cultivation of Musa spp lies with phytopathogenic fungi. Some fungi of great concern belong to the mycosphaerella-like group, since some 30 species have been described that directly affected plantain and banana leaves. However, only the following three species of the micosphaerella-like group cause considerable losses in these crops: Pseudocercospora eumusae (Mycosphaerella eumusae) is the cause of eumusae leaf spot; P. musae (M. musicola) causes yellow Sigatoka; and P. fijiensis (M. fijiensis) the black Sigatoka, the latter being the one of greatest concern worldwide.36 These 3 species directly affect the leaf of Musa spp., which alters the quality of the fruit. Regarding P. eumusae, it is found in India, Sri Lanka, Thailand, Malaysia, Vietnam, Mauritius, and Nigeria; however, it has not been found in the Americas.20 The yellow Sigatoka has been present in Puerto Rico since 1938-1939. This disease is characterized by yellow stripes in the leaves that later turn brown in the center. The described discoloring of the leaves affects the photosynthesis process that this organ carries out and, consequently, its adverse effect on the fruit. In the case of black Sigatoka, its presence in Puerto Rico has been identified since 2007.37 These are observed as reddish spots parallel to the venation of the leaves that later begin to turn a dark brown color. Then, the center of the spot begins to turn a grayish-white color, which ends up causing necrosis, affecting the production of chlorophyll. This can lead to a production loss of 85%.38 Other fungi that affect the production of plantain and banana are: 1) the cause of Cortana (Cordana musae), which is characterized by brown spots surrounded by yellow areas found on the margins of the leaf; 2) Cigar end rot, which is a rot at the tip of the fruit which causes many of these to mature prematurely. Among the fungi that cause this disease are: Stachylidium spp., Fusarium spp., and Deightoniella spp.33 All the fungi previously mentioned, except M. eumusae, have been observed in Puerto Rico. However, there is a fungal threat of primary concern to Musa spp. farmers worldwide and is the so called Panama Disease caused by Fusarium oxysporum.20
Fusarium oxysporum
Fusarium oxysporum is one of the fungal species of most significant phytopathological importance, the greatest number of host plants, and that causes the most critical economic damage among plant pathogens.39 Although pathogenic and non-pathogenic variants have been found in soil, this species can affect multiple plants of agricultural interest. This fungus mainly produces vascular wilting, followed by plant death. Some species can also cause crown and root rot. Fusarium oxysporum special forms (formae specialis) have caused significant epidemics in various crops such as banana, plantain, tomato, cotton, carnation, chrysanthemum, lentil, and chickpea, causing very substantial economic losses.40
The F. oxysporum special forms cannot be differentiated by their morphology or by the cultural characteristics of the colonies. However, they are physiologically different due to their ability to invade and cause diseases in specific host plants.41 This pathogenic selectivity of the special forms of F. oxysporum is because only the host plants and their radical exudates satisfy the nutritional requirements of the fungus, and, therefore, it can only grow and develop in these plants.42 The F. oxysporum special forms have a very dynamic genomic organization. Notable features of these and several species complexes within the genus Fusarium are chromosomal polymorphism43 and abundant transposable mobile elements, studied as possible contributors to differences in virulence.44 Further subdivisions of special forms in races are often made based on their virulence to a set of hosts of different cultivars, haplotypes, or varieties.45 Therefore, F. oxysporum is a very broad organism at the species level, with more than 120 different special forms that have been classified. Fusarium spp is part of the community of soil fungi and considered a natural component of the rhizosphere of plants: all strains of the species are saprophytic and can survive on organic matter, and some of them are phytopathogens that penetrate the roots of the plant host, inducing vascular wilt.46 Around 150 special forms and physiological races have been identified, including Foc F. oxysporum sp. cubense.
F. oxysporum sp. cubense
From here on, this review will focus on F. oxysporum sp. cubense (Foc), the special form that colonizes Musa spp. The main dispersal mechanisms of this pathogen are the movements of infected soil, runoff water, and the use of infected equipment on a farm (Figure 1).47 Since infected rhizomes are often symptomless, they effectively spread the pathogen when used as seeds. This fungus can survive for up to 30 years in the soil in the absence of banana due to its resistance structures called chlamydospores, making crop rotation ineffective in the short term.48 Furthermore, non-host weed species infected by the pathogen become inoculum reservoirs. Foc, a highly variable pathogen, comprises different evolutionary lineages.49 Somatic fusion and heterokaryon formation can occur between individuals regardless of sexual reproduction, but generally only between strains with similar genotypes.50 These unique networks of strains capable of heterokaryosis are termed vegetative compatible groups (VCG).51
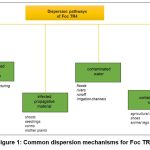 |
Figure 1: Common dispersion mechanisms for Foc TR4 |
The fungus is characterized by producing fast-growing colonies and three types of spores: microconidia, macroconidia, and chlamydospores. Microconidia are unicellular spores without septa, hyaline, ellipsoidal to cylindrical, straight, or curved. This fungus produces a white mycelium with a reddish background, producing elliptical or oval microconidia. Whereas macroconidia are thin-walled spores, spindle-shaped, long, moderately curved, with several cells and three to five transverse septa, with elongated basal cells and attenuated basal cells. While chlamydospores are spores formed by the transformation of cells of hyphae or macroconidia. They are characterized by having relatively thick walls, making them very resistant to unfavorable environmental conditions or the absence of host plants. Chlamydospores are formed singly or in pairs, are terminal or intercalary, and are mainly responsible for the survival of the fungus in dead tissues of host plants or soil.41 These chlamydospores can be present individually or in a chain that can last up to 30 years in the soil.52
The colonization of plants by Fusarium oxysporum f sp. cubense begins with its development in the root, in addition to access to the xylem. which does not always cause the disease.53,54 Once the plant recognizes the fungus, it produces antimicrobials and proteins that have to do with the pathogenesis process.55,56 Additionally, tyloses and other pectic binders are produced that help block the xylem vessels to prevent the spread of the fungus.57 This response passes between incompatible and compatible interactions, but the reaction seems to be very slow to prevent systemic infection in the latter. The first internal symptom seen is a reddish-brown discoloration of the xylem, which occurs in the feeder roots. This vascular discoloration extends to the rhizome and is largest where the stele meets the bark and continues to include large portions of the pseudostem. In plants four months old or older, symptoms can be seen on older leaves, such as yellowed leaves or longitudinally split leaves at the base.
Also the youngest leaves can wither and collapse until the entire canopy is made up of dead leaves or in that process.58 The growth of hyphae through the vascular vessels helps achieve rapid upward movement.59 The Xylem deterioration will limit or reduce water flow through the vessels, causing the wilting symptoms observed in infected plants. At the proteome level, “effector” proteins have been detected that help colonization with suppression or protection against the plant’s immune system.60
Banana root exudates help stimulate the germination of F. oxysporum f sp. cubense chlamydospores in the soil. The advance of the fungus through the roots is somewhat slow at first, but once it enters the pseudostem, it can spread very quickly through the formation of microconidia in the xylem vessels. Symptoms of infection may take two to six months to appear depending on the amount of initial inoculum, the environment and the resistance of the plant itself. In the last phase of the disease, the fungus moves from the xylem to the parenchyma and bark of the plant, where there are many chlamydospores and conidia that form in the degraded plant tissue.61 The virulence and epidemic nature of F. oxysporum f sp. cubense fall to its high pathogenicity and its enormous host range within the Musa genus. Different races of the pathogen are identified according to the pathogenicity in relation to the host cultivar. For instance, Race 1 (R1) affects Gros Michel (AAA) and Manzano / Apple / Latundan (Silk, AAB); Race 2 (R2) affects the Bluggoe’s cooking bananas (ABB); and Race 4 (R4) affects all cultivars in the Cavendish Subgroup (AAA) in addition to those susceptible to R1 and R2.62 Nevertheless, a population pathogen of Fusarium wilt in Heliconia spp was described as Race 3 but is no longer considered part of Foc.63
For Race 4, two variants have been identified: the tropical race (TR4), present in the tropical zone, and ST4, a variant present in the subtropical zone. These pathogen variants can infect bananas and plantains, as reported by Siamak and Zheng (2018).64 During the evaluation of the vegetative forms,58 concluded that the species associated with VCG1213, the causal agent of the current epidemic, belongs to a different species. This situation led to a new taxonomic classification for tropical race 4 and other species associated with races 1 and 2; therefore, since October 2018, TR4 has been known as F. odoratissimum, identified with the code FUSAC4.
In Latin America, TR4 is a quarantine pest due to the effects on bananas and plantains from Member Countries of the Andean Community (Bolivia, Colombia, Ecuador, and Peru), which require control of the pathogen to prevent spread.65 Several authors mention that it is essential to better understand where the pathogen is currently located and where it is expected to spread due to climate change. This way, they converted a database that includes the geodatabase and metadata about the 40 most important special forms of F. oxysporum, including FOC 4, into an interactive map.66-67 This map allows exploring the distribution of F. oxysporum incidence reports at the country level and its localities; as well as downloading essential metadata, and even submitting data from future studies to constantly update the web map and, in this way, serve as a long-term resource for the global Fusarium research community, as well as for governments as responsible for the policy formulation and other parties involved.
F. oxysporum races in Puerto Rico
Banana and plantain crops are the third most important in Puerto Rican agriculture after milk and poultry, according to USDA 2012 and 2017.68 However, opportunistic organisms such as Fusarium oxysporum f. sp. cubense (FOC) affect banana and plantain production. During the first decade of the twentieth century, a group of scientists found some symptomatic FOC characteristics after the rainy season.69 Later, during this decade, FOC race 2 was documented in the Mayagüez area.70 With the advent of technology, it was possible to isolate the organism and determine its genera using histologic observation techniques. However, these techniques did not permit the identification of the FOC race.71 Some studies in the late 1920s found FOC type 1 and FOC type 2 as the most common races present in Puerto Rico.
Further technological advances permitted the molecular identification to the level of species of Fusarium. For example, F. oxysporum f. sp. lycopersici affecting tomatoes was found from Puerto Rico to the U.S. south, including North Carolina, Arkansas, Georgia, and Florida.72 In addition, FOC race one was found in the north of the Island, in an Isabela banana plantation.16 However, there is still work to do to see FOC distribution in Puerto Rico using molecular approaches such as Next Generation Sequencing and phylogenetic methodologies.
Fusarium biotechnology
Identification and detection methodologies
The identification of Fusarium spp based on morphological and molecular characteristics, and the use of Koch’s postulates, are methodologies that lead to the identification of causal agents of diseases in plants. The morphological characterization of F. oxysporum is based on the macroconidia’s shape, the microconidiophores structure, and the chlamydospores formation and arrangement.73 Cultures are grown in nutritive media such as 1.5% agar-potato-dextrose (PDA) or Komada media for morphological characterization. A small piece of mycelium of approximately 5 mm2 is placed in each plate from a monosporic isolate of the fungus. The dishes are placed in an incubation chamber at a temperature of 22 °C for 8 days. Asexual reproduction in F. oxysporum is achieved by macroconidia and microconidia, whereas a sexual state of the fungus has never been observed.74
Pathogenic and nonpathogenic F. oxysporum species cannot be distinguished by morphology alone. It becomes necessary to perform pathogenicity tests. Pathogenic isolates of F. oxysporum exhibit a high level of host specificity directly related to its pathogenicity for various plant species.75 Therefore, the verification of Koch’s postulates is done by the inoculation of the pathogen in seedlings from several axenic cultures. Two inoculation methods with wounds and without wounds are implemented. Furthermore, absolute controls without wounds or inoculum are kept.
Molecular diagnostic studies of different banana growing areas found variants among the races of Foc, for which 20 compatible VCGs were found. DNA fingerprinting analyzes established genetic variation between compatible vegetative groups and physiological races. This pattern was specific for the VCG groups and showed evolution in four lineages, suggesting their evolution in Asia and multiple other areas.76 In general, mitochondrial DNA (mtDNA) RFLP patterns are identical within a VCG but vary between different VCGs in the same specific ways. VCG breed diversity is supported by genomic and mtDNA, and Restriction Fragment Profiling Length Polymorphism (RFLP). VCG and compatibility trials can identify TR4;77 however, these are time-consuming techniques.78
Researchers proposed a classification system for F. oxysporum strains based on their vegetative compatibility, and described a method based on nitrate pairing that does not use mutants to determine the VCG of each strain. At least 24 vegetative compatibility groups (VCG) are known to date in Foc,51,79 which can affect M. acuminata, M. balbisiana, M. schizocarpa, and M. textilis (Musaceae: Zingiberales).80 Isolates that are vegetatively compatible form a VCG group and typically share common biological, physiological, and pathological traits. Of the VCGs, 21 have been characterized, with most groups present in Asia, where the pathogen is believed to have evolved.81 The Foc-TR4 isolates are designated as VCG 01213 (or VCG 01216, a different designation for the same VCG). The Foc-STR4 isolates are designated as VCG 0120, 0121, 0122, 0129, and 01211.76 Since their emergence in Southeast Asia in the 1990s, they have caused severe damage to Cavendish plantations in Malaysia, Indonesia, China, the Philippines, the Northern Territory and Queensland in Australia, and Mozambique.82,83 Various DNA-based methods have been developed and tested for detecting Fusarium spp. associated with the disease. Most have focused on rapid identification of race 4, including VCGs associated with tropical and subtropical races. The primer group most used are the primers W2987 F W2987 R reported by Li et al. (2013), which encode a hypothetical protein that allows the discrimination of some genotypes associated with Foc.84
Other researchers developed a conventional PCR primer set capable of specifically detecting TR4 (VCG 01213) based on two SNPs in the IGS region. This primer set, Foc R4T F and R4T R, can differentiate between SR4 and TR4 more reliably than TEF-1a based primers. When multiplexed with appropriate primers, the IGS-based primer set can detect the pathogen in soil and the plant.85
The development of a real-time PCR test to detect VCGs 01213/16 and 0121 was based on a virulence gene previously described. The test is optimal for detecting the fungus in plant material, and evaluating reproducibility and specificity parameters, making it a useful test for rapidly identifying the fungus. This PCR test has been included as an initial part of a pre-diagnosis in pre-symptomatic and symptomatic plants. It has been considered as part of the diagnosis in a large number of reports and scientific articles, including the most re-cent incursions of the Tropical Race 4.86
The pre-diagnosis test requires isolating DNA samples from several suspected sources: symptomatic plants and soil. To isolate the fungus present in the plant, the PDA medium is used for plant sample cultivation, and the Komada medium is used for the soil sample using the methodology described by Garcia-Bastidas et al. (2020) in the Andean Guide for Fusarium oxysporum race 4 diagnoses. Once a monoculture sample is obtained, it can be used for DNA extraction and PCR tests.87
Comparative genomic studies have revealed that the host-specific pathogenicity of the F. oxysporum complex species (FOSC) was determined by different sets of supernumerary chromosomes (S.P.). In contrast to common vertical transfer, where genetic materials are transmitted through dividing cells, S.P. chromosomes can be transmitted horizontally between phylogenetic lineages, which explains the polyphyletic nature of the host-specific pathogenicity of FOSC.
With the increased understanding of gene interactions between Foc and bananas, future approaches for detecting specific pathotypes within the species complex could involve host-specific virulence factors, such as effectors. Such an approach enabled discrimination between different formae speciale of F. oxysporum tomato pathogens.88,89
To establish the presence of Fusarium race 4, the following actions are required to carry out the official report of the organism’s presence or new investment in a TR4-free country. The first stage is a molecular pre-diagnosis complemented with single spore tests if the samples taken from the plant are positive. The second stage requires sequencing, phylogenetic analysis, or the vegetative compatibility test (VCG3). This second stage requires a comparative analysis with the data obtained from countries previously reported with the presence of Race 4. Based on positive results from these previous stages, the third stage is carried out, which is the pathogenicity test, to verify the virulence of the isolated strains and complete Koch’s postulates. All these tests will confirm the presence of race 4 in a country.90
Efforts to create tolerance or resistance
Testing of hundreds of banana cultivars in Indonesia, many totally or partially resistant to Fusarium wilt, found that wild M. acuminata varieties were resistant.58 Also, the wild M. acuminata var. malaccensis from the Malaysian Peninsula91 or Sumatra were experimentally resistant to the disease. They argue that Indonesia is the primary gene center of Foc; therefore, it is likely to find a variety of disease resistance markers for the disease in bananas. Conventional breeding in bananas is difficult because of the extremely rare pro-duction of seeds. However, its parthenocarpy reduces the possibility of cross fertilization, making it an excellent candidate for genetic modification. The wild bananas identified as resistant to TR4 are appropriate candidates for gene mining.89 Several groups have identified such genes.92
A possible methodology to generate diversity and test for resistant/tolerant phenotypes is to induce mutations during breeding.93 In addition, collaborative efforts have been established to evaluate banana cultivars, such as the International Musa Testing Program coordinated by Bioversity International.92 For example, using prolonged multiplication in tissue culture and meticulous field screening, generated TR4-resistant Cavendish somaclones.89,94 Unfortunately, tissue culture variants selected for TR4 resistance (e.g., Cavendish-derived GCTCV genotypes) are not more resistant to other diseases than other varieties of Cavendish.95 However, these TR4-resistant varieties could become necessary if TR4 arrived in Puerto Rico.92
Formosana, a TR4-resistant variety of Giant Cavendish, has been developed in Taiwan. This variety, developed from somaclonal variation, has the same horticultural characteristics as Giant Cavendish but with moderate resistance to TR4. This variety has been approved for commercialization around the world. To our knowledge, Formosana has not been characterized in terms of its resistance mechanisms to TR4.96 Resistance genes could come from foreign sources, and some have conferred resistance in greenhouse trials. Paul et al. (2011) introduced an anti-apoptosis gene, a fungal-derived, and defense genes.97,98,99 A Cavendish line transformed with Ced9, an anti-apoptosis gene from C. elegans, demonstrated TR4 field resistance.100 The determination of their agronomic quality is pending.89
Some studies have developed transgenic Cavendish lines resistant to TR4.101 Three-year field resistance was demonstrated in one line transformed with the resistance gene RGA2 from a TR4-resistant diploid banana.100 Others are developing TR4-resistant Cavendish bananas by traditional efforts.102 TR4-resistant Cavendish (such as GCTCV-218) are planted in TR4-infested places, even if the cycling time is com-promised.89 In addition, efforts are underway to generate nontransgenic bananas us-ing CRISPR-Cas9.100 Ultimately, acceptance of these Cavendish cultivars depends on growers, traders, and consumers.89 The best control alternative is banana resistance to Fusarium. However, conventional screening processes are costly and time-consuming. Therefore, plants have evolved resistance proteins to detect effectors, which can be used as biological probes to reduce cost and time.89,103 This information about resistance proteins and the genetic variability in the island has led our research group to explore the possibility of developing plantains resistant to TR4.
Field analysis for resistance against Foc R4T
Foc TR4 was first detected in Australia in June 1997 initiating an immediate biosecurity response. However, more areas were found infected with Foc R4T and containment of the pathogen failed. Since then there has been a decline in the banana industry in the Northern Territory of Australia, and by 2012, the pathogen was declared endemic in this area; This part of the country being the only place in Australia where you can work with TR4. Since 2016, experiments began including reference banana varieties such as ‘Williams’ (very susceptible), ‘GCTCV 218’ (medium susceptible), ‘FHIA 01’ (resistant) and ‘FHIA 25’ (highly resistant). The evaluations in this trial were on external and internal symptoms in banana plants during harvest and during death, in addition to agronomic evaluations. After the trial, a disease severity score was assigned to each tested variety that ranged from 0 to 2, where 0 means that banana plants do not show disease symptoms under high inoculum pressure, and 2 means that the symptoms of the disease are serious, in addition to presenting high mortality rates due to the disease. As a result, more than 70% of the plants were affected . The first period of variety analysis was from June 2016 to 2018, in which all trials were artificially infected with approximately 200 ml of Foc R4T inoculum. Specifically, 24 banana varieties were evaluated, which were predominantly ‘Cavendish’ (‘GCTCV 106’, ‘GCTCV 215’, ‘GCTCV 247’, ‘GCTCV 218’, ‘CJ19’), FHIA hybrids (‘FHIA-18’ ‘ and ‘FHIA-25’) and parental lines (‘SH-3436’, ‘SH-3656’, ‘SH-3748’ and ‘SH-3217’). The plants were evaluated during two growing cycles that ended in March 2018. The results of this experiment show that 50% of the banana plants tested are resistant. Furthermore, some of the Chinese Cavendish varieties analyzed were rated as highly resistant.104 Table 1 summarizes some important data and characteristics of these varieties.
Table 1: Variants that have shown resistance in the field against Foc RT4.
| Cultivar | Origin | Characteristics | Reference |
| FHIA-25 | FHIA
Honduras |
Musa acuminata x
Balbisiana (AAB) |
Vézina (2019)105
Walduck and Daly(2017)106 Mintoff et al (2021)104 |
| FHIA-01 | FHIA
Honduras |
Musa acuminata × balbisiana Pome hybrid(AAAB) | Martínez- Solórzano et al (2020)107
Mintoff et al (2021)104 |
| GCTCV 215 | TBRI
Taiwan |
Cavendish
(AAA) |
Mintoff et al (2021)104 |
| FHIA-02 | FHIA
Honduras |
Williams(AAA) x SH-3393
Dessert hybrid (AAAA) |
Mintoff et al (2021)104 |
| SH-3362 (AT) | FHIA
Honduras |
Elite parent
(AA) |
Mintoff et al (2021)104 |
| SH-3362 | FHIA
Honduras |
Cooking hybrid
(AAB) |
Mintoff et al (2021)104 |
| Pisang Gajih Merah | Indonesia | ABB Saba | Mintoff et al (2021)104 |
| SH-3142 | FHIA
Honduras |
Elite parent
(AA) |
Mintoff et al (2021)104 |
| SH-3748 | FHIA
Honduras |
Cooking hybrid
(AAB) |
Mintoff et al (2021)104 |
| SH-3641 | FHIA
Honduras |
Pome hybrid
(AAAB) |
Mintoff et al (2021)104 |
| CJ19 | Indonesia | Cavendish
(AAA) |
Mintoff et al (2021)104 |
| FHIA-18 | FHIA
Honduras |
Prata Ana (AAB, Pome) x SH-3142
Pome hybrid (AAAB) |
Mintoff et al (2021)104 |
| GCTCV 247 | TBRI
Taiwán |
Cavendish
(AAA) |
Mintoff et al (2021)104
FAO (2022)108 |
Efforts to prevent Fusarium TR4 world wide
Fusarium threat and preventive measures
It has been recognized world-wide that the best option to avoid the impact of Foc TR4 on the production of Musaceae in a country is the exclusion of entry. Once this pest invades an area, the phytosanitary measures needed to prevent the movement of the pathogen from infected areas to free areas are costly and require trained personnel.109,110 Therefore, biosecurity measures for collecting, handling, and transporting suspicious samples can be successful if they become part of the Integrated Management of Pests and Diseases strategies. In 2019, the Food and Agriculture Organization of the United Nations (FAO), in its Regional Office for Latin America and the Caribbean, implemented an emergency project within its Technical Cooperation Program (TCP) “Strengthening of regional capacities for surveillance, prevention, and management against the possible spread of Fusarium wilt of bananas, caused by tropical race 4 of Fusarium oxysporum f.sp. cubense (Foc TR4)” to help the countries of Latin America and Caribbean countries fight against the spread of Fusarium TR4. The project sought to support regional coordination to achieve uniform application of phytosanitary measures and improve management and surveillance systems for this pest. It focuses on developing a regional action plan for the prevention, monitoring, and possible response to Foc TR4, among many other regional and national actions.111
The European Union defines interventions as the rationale for combining biological, biotechnological, chemical, cultivation, or plant selection measures. Regarding chemical intervention, it should be limited to the minimum necessary to keep the pest population at lower levels and reduce risks to human health and the environment. Furthermore, the intervention must implement an eradication-confinement program and an alternative program of containment suppression. The European Food Safety Authority has recommended measures to achieve the exclusion, containment, eradication, suppression, and surveillance of pathogens so that their own spread can be avoided or delayed in some way, as well as lower the level of intensity of emerging strains new Fusarium oxysporum outbreaks, including local outbreaks and endemic diseases.112
Phytosanitary geospatial analysis
Phytosanitary geospatial analysis is a new tool that can help strengthen and direct preventive actions for the spread of Fusarium wilt of Musaceae. This tool acts as a complement to the technical and scientific type that leads to the strengthening of the risk analysis carried out by the different organizations. This analysis can be carried out at the regional and national levels for phytosanitary protection.
This study has been carried out for other diseases, such as huanglongbing in Colombia or Xylella fastidiosa in Mexico.113,114 In this scene, Mexican researchers carried out a study to characterize potential areas under some level of phytosanitary risk from Fusarium oxysporum, f. sp. cubense race 4 tropical, to identify possible scenarios of introduction and establishment of the pathogen on a global scale with emphasis on the Americas. Spatial patterns of Foc TR4 at the global and subcontinental (pantropical America) levels were modeled, integrating the epidemic and spatial components. They included the potential distribution (pathogen/host) and the potential for economic damage by spatial association of trade nodes (origin/destination). At the territorial level, five levels of phytosanitary risk were characterized. On a global scale, they determined the existence of four focal areas with conditions for the presence of the disease. In pantropical America, 26,598 km2 were determined to be at very high phytosanitary risk (>0.9), located to a greater extent in Ecuador, Brazil, Mexico, Guyana, Guatemala, Venezuela, Panama, and Colombia. On a subcontinental scale, in the pantropical region of America, they identified an area of 15.8 million km2 with a lower level of risk.115
As an example, a recent study reported the first detection of Foc TR4 in the French department of Mayotte. In September 2019, leaf yellowing and wilting symptoms were observed in individual plants of the Silk (cv. Kissoukari) and Bluggoe (cv. Baraboufaka).
The Mayotte islands are very close to other islands, where bananas are considered an essential food, this discovery describes a great threat to the production of this fruit and to the economy of the population that depends on this crop.116This situation could be extrapolated to Puerto Rico due to its geographical location in the Caribbean and the risk area in which it is located, as mentioned above, which is confirmed in another work where it is mentioned that the risk that represent Foc-TR4 is enormous in banana producing countries in America continent especially, in the south and the Caribbean due to the close proximity of these zones.117
Consideration of this type of approach would be important in the strategies needed to protect our plantain crops. No reports have been found where the government of Puerto Rico has considered this approach.
Government level
Governments in potentially affected areas have implemented the request for certificates for disease indexing and strengthening of border control to avoid imports of bananas and plant parts from countries in which Foc-TR4 has been reported. Furthermore, they have also emphasized the importance of strengthening surveillance for early detection of potential pathogen introductions at farm sites, establishing quarantine sectors and promoting awareness campaigns to inform producers of the severe threat that Foc TR4 represents. Additionally, managing and controlling the movement of visitors and vehicles to farms by stringent cleaning and disinfection protocols of agricultural machinery, equipment, and tools among neighboring farms.
In the last decades, the Puerto Rico Department of Agriculture has promoted agricultural strategies to improve crop protection and help farmers care for their land, yet always dependent on Federal Aid from the USA. These efforts towards building up environmentally responsible agriculture have always demanded much and created a financial burden for farmers. Additionally, Puerto Rico’s location in the Caribbean has always increased its vulnerability to severe weather conditions, namely hurricanes, known to cause devastation on the island. Nonetheless, the government and Puerto Rico’s population have made serious efforts to create awareness, especially among young people, of the vital role we can all play in working towards sustainable agriculture.118
As stated in this text, agricultural products such as bananas and their different varieties, specifically plantain and Cavendish bananas, have become a significant part of our tropical cuisine. Consequently, these crops are in great demand, and their disease-free harvest is a constant need and challenge. In 2019, the former secretary of the Department of Agriculture in Puerto Rico, the agronomist Carlos Flores-Ortega, expressed concerns to banana plant producers about the appearance of Fusarium sp. TR4 in Colombia. Flores-Ortega confirmed what other studies had also reported, namely, that the fungus TR4 did not exist on the Island, yet preventive measures should be outlined and followed. The former secretary also stated that bananas had not been imported for several years, and their consumption had been 100% from local farms.119
The same year, the government of Puerto Rico proceeded to announce and implement specific measures similar to those accepted worldwide to prevent FOC TR4. Among the phytosanitary measures were rigorous quarantine for imports of bananas to Puerto Rico; establishing a program, in coordination with the College of Agricultural Sciences of the Mayagüez Campus of the University of Puerto Rico, to educate and train agronomists; monitoring farms by regions and establishing biosafety controls.
The government of Puerto Rico, based on Law 76 for Emergency Situations or Events Procedures Act, gave legal support to Phytosanitary Emergencies for all measures to help cope with pests and apply regional sanitation standards. They demanded that deficiencies in compliance be identified and periodically evaluated. As of 2019 and still in 2023, no TR4 strains have been identified in Puerto Rico, yet the island is not exempt from a possible epidemic. There is hope that banana researchers (such as those working at the research center-CEIBA) can develop a resistant cultivar and the use of fungicides, such as Isotianil (Stout®), can induce systemic acquired resistance in plants 120. The use of diagnostics protocols for Foc TR4 should also be encouraged among farm personnel. The research group in CEIBA is also developing diagnostic protocols to meet those needs.
Conclusions
Due to its lethality, Panama disease, or Fusarium wilt, is the most important banana disease. It is present in all commercial areas of this crop worldwide on four continents, except Europe. The disease’s epidemic spread exacts immense economic tolls, amounting to millions of dollars for producing nations, owing to its severe repercussions. A framework of risk elements and criteria has been established to counteract this threat, aimed at preventing disease entry, establishment, spread, and impact. Consequently, enhancing the sensitivity of detection methods becomes pivotal to preempting disease dissemination within disease-free regions. Concurrently, reinforcing surveillance endeavors in at-risk areas necessitates advanced tools that align with scientific and technological advancements. Puerto Rico, a densely populated small island, struggles with challenges concerning food security. Within this context, bananas and plantains emerge as paramount local food crops, amplifying the need for effective disease management. In Puerto Rico’s unique circumstances, a comprehensive review of local research on Musa spp. and associated pathogens takes on vital importance. The island’s compact area houses a multitude of physical landscapes, fostering continuous research endeavors that yield substantial insights. Nonetheless, the island’s diverse environments are dual, bringing challenges and opportunities for developing disease-resistant Musa cultivars. Crafting cultivars capable of adapting to the island’s multiple physical settings, containing test pathogens, and mitigating crop losses from tempestuous winds represents an intricate puzzle. Conversely, the island’s proximity to diverse environments and genetic variability provides a fertile ground for cultivating pathogen-resistant varieties.
Acknowledgment
The authors thank Dr. Fatima Gandarilla and Dr. Lumary Pérez-Guzmán for their invaluable edits and insights on our manuscript.
Conflict of Interests
The authors do not have any conflict of interest.
Funding Sources
This work was carried out with the support of our research center, Biotechnology and Agrobiotechnology Research and Learning Center, and our university, Pontifical Catholic University of Puerto Rico.
References
- Álvarez E., Pantoja A., Gañan L. y Ceballos G. Current Status of Moko Disease and Black Sigatoka in Latin America and the Caribbean, and options for managing them. Cali, Colombia: Centro Internacional de Agricultura Tropical; 2013. pp.40
- Esteban- Guapacha S., Salazar- Salazar M., Aguillón J. and Landázuri P. Karyotypic similarity among different cultivars of Musa spp. from Quindío-Colombia. Cultivos Tropicales. 2017; 38:119-126.
- Regional International Organization for Agricultural Health. Risk analysis Fusarium oxysporum f. sp. cubense race 4 tropical (Foc R4T) Quarantine pest. F&G Editores, San Salvador., El Salvador, 2019; pp.287
- García-Velasco R., Portal-González N., Santos-Bermúdez R., Yanes-Paz E., Lorenzo-Feijoo J.C. y Companioni-González B. Rapid method applied in previous evaluation of banana resistance to Fusarium oxysporum f. sp. cubense. Rev. Mex. Fitopatol. 2020; 38(3): 384-397. DOI: 10.18781/r.mex.fit.2004-1.
CrossRef - Food and Agriculture Organization of the Nations United. https://www.fao.org/3/y5102s/ y5102s03.htm#TopOfPage(accessed on 30 july 2023).
- Spanish Nutrition Foundation. Banana. https://www.fen.org.es/MercadoAlimentosFEN/ pdfs/platano.pdf(accessed on 30 july 2023).
- Food and Agriculture Organization of the Nations United. Medium-term outlook: prospects for global production and trade of bananas and tropical fruits 2019-2028.https://www.fao.org/ documents/ card/en/c/CA7568ES (accessed on 22 july 2023).
- Food and Agriculture Organization of the Nations United. Markets and Trade. Bananas. https://www.fao.org/markets-and-trade/commodities/bananos/es/(accessed on 18 july 2023).
- FAOSTAT 2023. Food and Agriculture Organization of the Nations United https://www.fao.org/faostat/en/(accessed on 22 july 2023).
- Irish B., Rios C., Daniells J. and Goenaga R. Catalog of banana (Musa spp.) accessions maintained at the USDA-ARS Tropical Agriculture Research Station, USDA-ARS, United States of America;2016. pp.1-296.
- Pereira A. and Maraschin M. Banana (Musa spp) from peel to pulp: ethnopharmacology, source of bioactive compounds and its relevance for human health. J. Ethnopharmacol. 2015;160 (3)149-163. DOI: 10.1016/j.jep.2014.11.008.
CrossRef - Santos A.S., Amorim E.P., Ferreira C.F. and Pirovani C.P. Water stress in Musa spp.: A systematic review. PLoS One. 2018; 12 e0208052. DOI: 10.1371/journal.pone.0208052.
CrossRef - Nadal-Medina R., Manzo-Sánchez G., Orozco-Romero J., Orozco-Santos M. y Guzmán-González S. Genetic diversity of bananas and plantains (Musa spp.) determined using RAPD markers. Rev. Fitotec. Mex. 2009; 32(1) 01-07.
CrossRef - Nelson S.C., Ploetz R. C. and Kepler A. K. Musa species (banana and plantain). Species profiles for Pacific Island agroforestry. 2006; 15:251-259.
- Perrier X., De Langhe E., Donohue M., Lentfer C., Vrydaghs L., Bakry F., Carreel F., Hippolyte I., Horry J.P., Jenny C., Lebot V., Risterucci A.M., Tomekpe K., Doutrelepont H., Ball T., Manwaring J., de Maret P. and Denham, T. Multidisciplinary perspectives on banana (Musa spp.) domestication. PNAS. 2011; 108(28): 11311-11318. DOI:10.1073/pnas.1102001108.
CrossRef - Garcia R.O., Rivera-Vargas L.I., Ploetz R., Correll J. C. and Irish B. M. Characterization of Fusarium spp. isolates recovered from bananas (Musa spp.) affected by Fusarium wilt in Puerto Rico. Eur. J. Plant Pathol. 2018; 152:599-611. DOI:10.1007/s10658-018-1503-y.
CrossRef - USDA NASS (2020) 2017 Census of Agriculture: Puerto Rico (2018) Island and Regional Data. Volume 1. Geographic Area Series Part 52. AC-17-A-52.
- Irish B.M., Crespo A., Goenaga R., Niedz R. and Ayala-Silva T. Ploidy level and genomic composition of Musa spp. accessions at the USDA-ARS Tropical Agriculture Research Station. The Journal of Agriculture of the University of Puerto Rico. 2009; 93 (1-2):1-21. DOI:10.46429/jaupr.v93i1-2.2750.
CrossRef - Irish B., Goenaga R., Rios C., Chavarria-Carvajal J. and Ploetz R. Evaluation of banana hybrids for tolerance to black leaf streak (Mycosphaerella fijiensis Morelet) in Puerto Rico. Crop Protection.2013; 54:229-238. DOI:10.1016/j.cropro.2013.09.003.
CrossRef - Manzo-Sánchez G., Orozco-Santos M., Martínez-Bolaños L., Garrido-Ramírez E. y Canto-Canche B. Diseases of quarantine and economic importance of banana (Musa sp.) cultivation in Mexico. Rev. Mex. Fitopatol. 2014;32(2): 89-107.
- Brown A., Tumuhimbise R., Amah D., Uwimana B., Nyine M., Mduma H., Talengera D., Karamura D., Kuriba J. and Swennen R. Bananas and plantains (Musa spp.). In: Campos H., Caligari P.D.S., Eds. Genetic improvement of tropical crops. Berlin, Germany: Springer International Publishing; 2017: pp. 219-240.
CrossRef - Kumar P. L., Selvarajan R., Iskra-Caruana M.L., Chabannes M. and Hanna, R. Biology, etiology, and control of virus diseases of banana and plantain. Adv. Virus Res. 2015; 91:229-269. DOI:10.1016/bs.aivir.2014.10.006.
CrossRef - Waghmare S. S., Adat S. R., Mohite V. K., Waghule A.A. and Patale S.S. Study of bunchy top of banana virus (BBTV) and its control by integrated disease management. Int. J. Curr. Microbiol. App. Sci. 2021;10(10): 416-429. DOI: 10.20546/ijcmas.2021.1010.047.
CrossRef - Hu J., Xu M., Wu Z. and Wang M. Detection of banana bunchy top virus in Hawaii. Plant Dis. 1993; 77:952. DOI: 10.1094/PD-77-0952D.
CrossRef - Martinez R. T., Renjifo D., Cayetano X., Pineau K. P., Umber M. and Teycheney P.Y. Prevalence and diversity of Banana streak viruses in the Dominican Republic. Trop. Plant Pathol. 2020; 45:376-384. DOI:10.1007/s40858-020-00334-z.
CrossRef - Kolombia Y., Oviasuyi T., Ayisah K. D., Ale Gonh-Goh A., Atsu T., Oresanya A., Ogunsanya P., Alabi T. and Kumar P. First report of banana bunchy top virus in banana (Musa spp) and its eradication in Togo. Plant Dis. 2021;105(10):3312. DOI: 10.1094/PDIS-03-21-0473-PDN.
CrossRef - Jooste A., Wessels N. and Van der Merwe M. First Report of Banana bunchy top virus in Banana (Musa spp.) from South Africa. Plant Dis. 2016;100 (6):1251. DOI: 10.1094/PDIS-12-15-1422-PDN.
CrossRef - Lepcha S., Chaudhary K. and Pratap D. First report of Cucumber mosaic virus infecting Musa × paradisiaca cv. Chini champa in Sikkim, Northeast India. Plant Dis. 2017;101 (5):844-844. DOI:10.1094/PDIS-07-16-1081-PDN.
CrossRef - Selvarajan R., Balasubramanian V. and Gayathrie T. Highly efficient immunodiagnosis of episomal banana streak my virus using polyclonal antibodies raised against recombinant viral-associated protein. J Phytopathol. 2016; 164(7-8) 497-508. DOI:10.1111/jph.12475
CrossRef - Selvarajan R., Balasubramanian V., Jeyabaskaran K. J., and Mustaffa M.M. Yield loss management through fertilizers in Banana bract mosaic virus affected French plantain cv. Nendran (AAB). Indian J Agr. Sci. 2017; 87 (8):1055-1061.
CrossRef - Ramírez J. G., Muñoz M., Patiño L. F. and Morales J.G. Banana Moko disease management with resistance inducers and chlorine dioxide. Agronomía Colombiana. 2015; 33 (2):194-202. DOI:10.15446/agron.colomb.v33n2.48663.
CrossRef - Tripathi L., Mwangi M., Abele S., Aritua V., Tushemereirwe W. K. and Bandyopadhyay R. Xanthomonas wilt; a threat to banana production in East and Central Africa. Plant Dis.2009; 93 (5): 440-445. DOI:10.1094/ PDIS-93-5-0440.
CrossRef - Alvarado-Ortiz A.N. y Díaz M. Practical guide to pests and diseases in plantains and bananas. Puerto Rico: Agricultural Extension Service. College of Agricultural Sciences. Mayagüez University Campus;2007. pp.15-18.
- Rengifo J., Zapata M., Díaz M. and Ingles R. Pathogenicity of bacteria isolated from ’Hua Moa’ in with bunch abortion symptoms inoculated into other clones of plantain and banana. The Journal of Agriculture of the University of Puerto Rico. 2007; 91(1-2): 19-30. DOI:10.46429/jaupr.v91i1-2.2650.
CrossRef - Fulton J., Bec S., Fayette J., Ploetz R., Garrett K.A. and Harmon C. L. First report of plantain soft rot caused by Klebsiella variicola in Haiti. 2020; Plant Dis.104(6): 1851. DOI:10.1094/PDIS-10-19-2105-PDN.
CrossRef - Crous P., Carlier J., Roussel V. and Groenewald J. Pseudocercospora and allied genera associated with leaf spots of banana (Musa spp.). Fungal Systematics and Evolution. 2021; 7:1-19. DOI: 10.3114/fuse.2021.07.01.
CrossRef - Irish B.M., Goenaga R. and Ploetz R.C. Mycosphaerella fijiensis, causal agent of black Sigatoka of Musa spp. found in Puerto Rico and identified by polymerase chain reaction. Plant Dis. 2006; 90 (5): 684. DOI: 10.1094/PD-90-0684A.
CrossRef - Ugarte- Fajardo J., Bayona- Andrade O., Criollo- Bonilla R., Cevallos-Cevallos J., Mariduena-Zavala M., Ochoa Donoso D. and Vicente Villardón J.L. Early detection of black Sigatoka in banana leaves using hyperspectral images. Appl. Plant Sci. 2020; 28(8):e11383. DOI: 10.1002/aps3.11383.
CrossRef - Cai H., Yu N., Liu Y., Wei X. and Guo C. Meta-analysis of fungal plant pathogen Fusarium oxysporum infection-related gene profiles using transcriptome datasets. Front. Microbiol. 2022; 13:970477. DOI: 10.3389/fmicb.2022.970477.
CrossRef - Edel-Hermann V. and Lecomte, C. Current Status of Fusarium oxysporum Formae Speciales and Races. Phytopathology. 2019;109 (4):512-530. DOI:10.1094/PHYTO-08-18-0320-RVW.
CrossRef - Nelson P.E., Horst R.K. and Woltz S.S. Fusarium Diseases of Ornamental Plants. In Nelson P.E., Toussoun T.A. and Cook R.J., Eds. Fusarium: Diseases, Biology and Taxonomy. University Park, PA, and London: The Pennsylvania State University Press; 1981: pp. 121-128.
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology.2005;105 (12):1512-1521. DOI:10.1094/PHYTO-04-15-0101-RVW.
CrossRef - Boehm E. W. A., Ploetz R. C. and Kistler H.C. Statistical analysis of electrophoretic karyotype variation among vegetative compatibility groups of Fusarium oxysporum f.sp. cubense. Mol. Plant Microbe In. 1994; 7:196-207. DOI : 10.1094/MPMI-7-0196.
CrossRef - Daboussi M.J. and Capy P. Transposable elements in filamentous fungi. Ann. Rev. Microbiol. 2003; 57:275-299. DOI :10.1146/annurev.micro.57.030502.091029.
CrossRef - Pires da Silva F., Vechiato M. and Harakava R. EF-1α gene and IGS rDNA sequencing of Fusarium oxysporum f. sp. vasinfectrum and F. oxysporum f. sp. phaseoli reveals polyphyletic origin of strains. Trop. Plant Pathol. 2014;39(1):64-73. DOI:10.1590/S1982-56762014000100008.
CrossRef - López-Zapata S.P. and Castaño-Zapata J. Integrated management of Panama disease Fusarium oxysporum Schlechtend.: Fr. sp. cubense (E.F. SM.) W.C. Snyder & H.N. Hansen: a review. Revista U.D.C.A Actualidad & Divulgación Científica. 2019;22: e1240. DOI:10.31910/rudca.v22.n2.2019.1240.
CrossRef
- Dita M., Barquero M., Heck D., Mizubuti E. S. G. and Staver C.P. Fusarium Wilt of Banana: Current Knowledge on Epidemiology and Research Needs Toward Sustainable Disease Management. Front Plant Sci. 2018;9:1468. DOI: 10.3389/fpls.2018.01468.
CrossRef - Daugovish O., Smith R., Cahn M., Koike S., Smith H., Aguiar J., Quiros C., Cantwell M. and Takele E. Celery production in California. University of California, USA: The Regents Division of Agriculture and Natural Resources;2008. pp.4
CrossRef - O’Donnell, K., Gueidan C., Sink S., Johnston P. R., Crous P. W., Glenn A., Riley R., Zitomer N. C., Colyer P., Waalwijk C., van der Lee T., Moretti A., Kang S., Kim H.S., Geiser D. M., Juba J. H., Baayen R. P., Cromey M. G., Bithell S., Sutton D. A., Skovgaard K., Ploetz R., Kistler H. C.,Elliot M., Davis M. and Sarver B.A.J. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009; 46 (12):936-948. DOI: 10.1016/j.fgb.2009.08.006.
CrossRef - Kistler H.C. Genetic Diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology.1997; 87 (4):474-479. DOI:10.1094/PHYTO.1997.87.4.474.
CrossRef - Puhalla, J. E. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Can. J. Bot.1985;63 (2):179-183. DOI:10.1139/b85-020.
CrossRef - Ploetz R.C. Fusarium Wilt of Banana Is Caused by Several Pathogens Referred to as Fusarium oxysporum f. sp. cubense. Phytopathology. 2006;96 (6):653-656. DOI:10.1094/PHYTO-96-0653.
CrossRef - Jiménez-Fernández D., Landa B.B., Kang S., Jiménez-Díaz R.M. and Navas-Cortés J.A. Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris Races. PLOS ONE.2013; 8(4): e61360. DOI: 10.1371/journal.pone.0061360.
CrossRef - Gordon T. R. Fusarium oxysporum and the Fusarium Wilt Syndrome. Ann. Rev. Phytopathol. 2017; 55:23-39. DOI:10.1146/annurev-phyto-080615-095919.
CrossRef - Rep M., Meijer M., Houterman P.M., Van Der Does H.C. and Cornelissen B.J.C. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant Microbe Interact. 2005;18 (1):15-23. DOI:10.1094/MPMI-18-0015.
CrossRef - dos Santos C. and Franco O.L. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants. 2023;12 (11) 2226. DOI:10.3390/plants12112226.
CrossRef - Kashyap A., Planas-Marquès M., Capellades M., Valls M. and Coll N. S. Blocking intruders: inducible physico-chemical barriers against plant vascular wilt pathogens. J. Exp. Bot. 2021;72 (2): 184-198. DOI: 10.1093/jxb/eraa444.
CrossRef - Maryani N., Lombard L., Poerba Y., Subandiyah S., Crous P. and Kema G. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019;92:155-194. DOI: 10.1016/j.simyco.2018.06.003.
CrossRef - Michielse C.B. and Rep M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009;10 (3):311-324. DOI: 10.1111/j.1364-3703.2009.00538.
CrossRef - van Dam P., Fokkens L., Ayukawa Y., van der Gragt M., ter Horst A., Brankovics B., Houterman P.M., Arie T. and Rep M. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 2017; 7(1): 9042 DOI:10.1038/s41598-017-07995-y.
CrossRef - Pegg K. G., Coates L. M., O’Neill W. T. and Turner D.W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 2019; 10:1395. DOI: 10.3389/fpls.2019.01395.
CrossRef - Su H., Hwang S. and Ko W. Fusarial wilt of Cavendish bananas in Taiwan. Plant Dis. 1986;70:814-818. DOI: 10.1094/PD-70-814.
CrossRef - Ploetz, R.C. Panama disease, an old nemesis rears its ugly head: Part 1, the beginnings of the banana export trades. Plant Health Progress.2005;6 (1):1-11. DOI: 10.1094/PHP-2005-1221-01-RV.
CrossRef - Siamak S.B. and Zheng S. Banana Fusarium wilt (Fusarium oxysporum f. sp. cubense) control and resistance, in the context of developing wilt-resistant bananas within sustainable production systems. Horticultural Plant Journal. 2018;4 (5):208-218. DOI: 10.1016/j.hpj.2018.08.001.
CrossRef - García-Bastidas F.A., van der Veen A.J.T., Nakasato-Tagami G., Meijer H.J.G., Arango-Isaza R.E. and Kema G.H.J. An Improved Phenotyping Protocol for Panama Disease in Banana. Front. Plant Sci. 2019; 10:1006. DOI: 10.3389/fpls.2019.01006
CrossRef - Calderón R., Eller J. A., Brodsky H. K., Miles A. D., Crandall S. G., Mahowald N., Pavlick R. and Gold K. M. An Interactive, Online Web Map Resource of Global Fusarium oxysporum ff. spp. Diversity and Distribution. Plant Dis. 2023; 107:538-541. DOI: 10.1094/PDIS-04-22-0789-A.
CrossRef - Gold Lab Grape Sensing, Pathology, and Extension at Cornell – GrapeSPEC. Fusarium oxysporum webmap. https://blogs.cornell.edu/goldlab/fusarium-oxysporum-webmap/ (accessed on 22 july 2023).
- United States Departament of Agriculture. National Agricultural Statistics Service. https://www.nass.usda.gov/Publications/Highlights/index.php(accessed on 28 july 2023).
- Fawcett W. The banana: Its cultivation, distribution and commercial uses, 2 ed. London, England: Duckworth and Company;1913.
- Brandes E. W. Banana wilt. Phytopathology. 1919;9:339-389.
CrossRef - Leslie J. F. and Summerell B. A. The Fusarium laboratory manual, 1 ed. Iowa. USA,Blackwell Publishing;2006.
CrossRef - Gale L. R. Katan T. and Kistler H. The probable center of origin of Fusarium oxysporum f. sp. lycopersici VCG 0033. Plant Dis. 2003; 87 (12):1433-1438. DOI: 10.1094/PDIS.2003.87.12.1433.
CrossRef - Unda F., Agüero J., Fariñas M.C. and Martínez- Martínez L. Identification of fungi of clinical importance using molecular techniques. Infectious Diseases and Clinical Microbiology. 2011; 29:282-285. DOI:10.1016/j.eimc.2010.12.011.
CrossRef - Booth C. Fungal Culture Media. In Booth C., Eds. Methods in Microbiology. Volume 4. Great Britain: Academic Press:London; 1971: pp.49-94.
CrossRef - Fravel D., Olivain C. and Alabouvette C. Fusarium oxysporum and its biocontrol. New Phytol. 2003;157 (3):493-502. DOI: 10.1046/j.1469-8137.2003.00700.x
CrossRef - Bentley S., Pegg K.G., Moore N. Y., Davis R.D. and Buddenhagen I.W. Genetic variation among vegetative compatibility groups of Fusarium oxysporum f.sp. cubense analyzed by DNA fingerprinting. Phytopathology.1998;88 (12): 1283-1293. DOI:10.1094/PHYTO.1998.88.12.1283
CrossRef - Correll J.C., Klittich C.J.R. and Leslie J.F. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology. 1987; 77 (12):1640-1646.
CrossRef - Dita M. A., Waalwijk C., Buddenhagen I.W., Souza Jr M. T. and Kema G. H. J. A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen. Plant Pathol. 2010;59 (2):348-357 DOI: 10.1111/j.1365-3059.2009.02221.x.
CrossRef - Mostert D., Molina A.B., Daniells J., Fourie G., Hermanto C., Chao C., Fabregar E., Sinohin V. G., Masdek N., Thangavelu R., Li C., Yi G., Mostert L. and Viljoen A. The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE. 2017;12 (7): e0181630. DOI:10.1371/journal.pone.0181630
CrossRef - Ploetz R.C. Fusarium wilt of banana. Phytopathology. 2015;105 (12):1512-1521. DOI: 10.1094/PHYTO-04-15-0101-RVW.
CrossRef - Fourie G., Steenkamp E.T., Gordon T.R. and Viljoen A. Evolutionary relationships among the Fusarium oxysporum f.sp. cubense vegetative compatibility groups. Appl. Environ. Microbiol. 2009;75 (14):4770-4781. DOI: 10.1128/AEM.00370-09.
CrossRef - Koenig R. L., Ploetz, R. C. and H. C. Kistler. Fusarium oxysporum f. sp. cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathology.1997;87(9):915-923. DOI:10.1094/PHYTO.1997.87.9.915.
CrossRef - Buddenhagen, I.W. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of ‘tropical race 4’ to better manage banana production. Acta Hortic. 2009; 828:193-204. DOI: 10.17660/ActaHortic.2009.828.19
CrossRef - Li C., Shao J., Wang Y., Li W., Guo D., Yan B., Xia Y. and Peng M. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genomics. 2013c;14:851. DOI: 10.1186/1471-2164-14-851.
CrossRef - Dita M. A., Waalwijk C., Buddenhagen I.W., Souza Jr M. T. and Kema G. H. J. A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen. Plant Pathol. 2010;59 (2):348-357 DOI: 10.1111/j.1365-3059.2009.02221.x.
CrossRef - Lin Y.H., Su C.C., Chao C.P., Chen C.Y., Chang C.J., Huang J.W. and Chang P.F.L. A molecular diagnosis method using real-time PCR for quantification and detection of Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 2013; 135:395-405. DOI: 10.1007/s10658-012-0096-0.
CrossRef - García-Bastidas F. A., Quintero-Vargas J. C., Ayala-Vasquez M., Shermer T., Sedl M.F., Santos-Paiva M., Noguera A.M., Aguilera-Galvez C., Wittenberg R., Sorensen A. and Kema G.H. First Report of Fusarium Wilt Tropical Race 4 in Cavendish Bananas caused by Fusarium odoratissimum in Colombia. Plant Dis. 2020;104 (3): 994. DOI:10.1094/PDIS-09-19-1922-PDN
CrossRef - Lievens B., Houterman P.M. and Rep M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales, FEMS Microbiol. Lett. 2009;300 (2) 201-215, DOI: 10.1111/j.1574-6968.2009.01783.x
CrossRef - Viljoen A., Ma L.J. and Molina A.B. Fusarium wilt (Panama disease) and monoculture banana production: resurgence of a century-old disease. In Ristaino J.B. and Records A. Eds. Emerging Plant Diseases and Global Food Security. Minnesota, USA: APS Press; 2020: pp. 159-184.
CrossRef - Buddenhagen I.W. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of ‘tropical race 4’ to better manage banana production. Acta Hortic. 2009;828:193-204. DOI: 10.17660/ActaHortic.2009.828.19
CrossRef - Javed M., Chai M. and Othman R. Study of Resistance of Musa acuminata to Fusarium oxysporum using RAPD markers. Biol. Plantarum. 2004; 48: 93-99. DOI: 10.1023/B:BIOP.0000024281.85427.6d.
CrossRef - Irish B., Goenaga R., Montalvo-Katz S., Chaves-Cordoba B. and Van den Bergh, I. Host response to black leaf streak and agronomic performance of banana genotypes in Puerto Rico. Hort. Science. 2019;54 (10): 1808-1817. DOI: 10.21273/HORTSCI13876-19.
CrossRef - Ortiz R. and Swennen R. From crossbreeding to biotechnology-facilitated improvement of banana and plantain. Biotechnol. Adv. 2014;32 (1):158-169. DOI: 10.1016/j.biotechadv.2013.09.010.
CrossRef - Hwang S.C. and Ko W.H. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 2004;88 (6):580-588. DOI: 10.1094/PDIS.2004.88.6.580.
CrossRef - Molina A.B., Fabregar E.G., Soquita R.O. and Sinohin V.G.O. Comparison of host reaction to Fusarium oxysporum f. sp. cubense Tropical Race 4 and agronomic performance of somaclonal variant ‘GCTCV 119’ (AAA, Cavendish) and ‘Grand Naine’ (AAA, Cavendish) in commercial farms in the Philippines. Acta Hortic. 2011; 897:391-393. DOI:10.17660/ActaHortic.2011.897.55.
CrossRef - Taiwan Banana Research Institute. https://www.promusa.org/Taiwan+Banana+Research+Institute+-+TBRI (accessed on 28 july 2023).
- Paul J.Y., Becker D. K., Dickman M.B., Harding R. M., Khanna H, K. and Dale J.L. Apoptosis-related genes confer resistance to Fusarium wilt in transgenic ‘Lady Finger’ bananas. Plant Biotechnol. 2011;9 (9): 1141-1148. DOI:10.1111/j.1467-7652.2011.00639.x
CrossRef - Ghag S.B., Shekhawat U.K. and Ganapathi T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014a; 12 (5):541-53. DOI:10.1111/pbi.12158.
CrossRef - Ghag S.B., Shekhawat U.K. and Ganapathi T.R. Native cell-death genes as candidates for developing wilt resistance in transgenic banana plants. AoB Plants. 2014b;6:plu037. DOI:10.1093/aobpla/plu037.
CrossRef - Dale J., James A., Paul J.Y., Khanna H., Smith M.; Peraza-Echeverria S.; Garcia-Bastidas F., Kema G., Waterhouse P., Mengersen K. and Harding R. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017b; 8:1496. DOI:10.1038/s41467-017-01670-6.
CrossRef - Dale J., Paul J.Y., Dugdale B. and Harding R. Modifying Bananas: From Transgenics to Organics? Sustainability.2017a;9 (3):333. DOI:10.3390/su9030333.
CrossRef - Aguilar Morán J.F. Improvement of Cavendish Banana cultivars through conventional breeding. Acta Hortic. 2013;986: 205-208.DOI: 10.17660/ActaHortic.2013.986.2.
CrossRef - Maekawa T., Kufer T. A. and Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 2011; 12:818-826. DOI: 10.1038/ni.2083.
CrossRef - Mintoff S. J., Nguyen T. V., Kelly C., Cullen S., Hearnden M., Williams R. and Tran-Nguyen, L. T. 2021. Banana Cultivar Field Screening for Resistance to Fusarium oxysporum f. sp. cubense Tropical Race 4 in the Northern Territory. J. Fungi. 7(8): 627.
CrossRef - Vézina, A. 2019. Tropical race 4. ProMusa, Rome, ITA. http://www.promusa.org/Tropical+race+4+-+TR4). (accessed on 22 october 2023).
- Walduck G. and Daly A. Identification of Banana Varieties with Resistance to Fusarium wilt Tropical Race 4; Report to Horticulture Australia Limited, Project No. FR00043: Hort Innovation Australia: North Sydney, NSW, Australia; 2007.
- Martínez-Solórzano G. E., Rey-Brina J. C., Pargas-Pichardo R. E. y Manzanilla E. E. Fusarium wilt tropical race 4: Current status and presence in the American continent. Agronomía Mesoamericana. 2020; 31(1):259-276.
CrossRef - FAO 2022. Web Seminar Banana varieties resistant to Foc R4T: from selection to market demand. Global Foc R4T Network Report 2022. https: //www.fao.org/3/cb9345es/cb9345es.pdf (accessed on 22 october 2023).
- Olivares-Barlin O., Rey- Juan C., Lobo D., Navas-Cortés J.A., Gómez J. A. and Landa- Blanca B. Fusarium Wilt of Bananas: A Review of Agro-Environmental Factors in the Venezuelan Production System Affecting Its Development. Agronomy .2021;11(5): 986. DOI: 10.3390/agronomy11050986.
CrossRef - CABI. Invasive Species Compendium. Wallingford, UK: CAB International. https://www.cabidigitallibrary.org/doi/full/10.1079/cabicompendium.24621(accessed on 30 july 2023).
- Food and Agriculture Organization of the Nations United. TR4 Global Network https://www.fao.org/tr4gn/fao-in-action/en/ (accessed on 16 july 2023).
- Bragard C., Baptista P., Chatzivassiliou E., Di Serio F., Gonthier P., Jaques-Miret J.A., Justesen A.F., MacLeod A., Magnusson C.S., Milonas P., Navas-Cortes J.A., Parnell S., Potting R., Stefani E., Thulke H.H., Van der Werf W., Civera A.V., Yuen J., Zappala L., Migheli Q., Vloutoglou I., Maiorano A., Streissl F. and Reignault P.L. 2022. Scientific Opinion on the pest categorization of Fusarium oxysporum f. sp. cubense Tropical Race 4. EFSA Journal. 2022;20 (1):7092. DOI:10.2903/j.efsa.2022.7092.
CrossRef - Olvera-Vargas L. A., Quiroz -Gaspar A.J., Contreras-Medina D. I. y Aguilar-Rivera N. Huanglongbing potential risk analysis through geospatial technology in Colombia. Ciencia y Tecnología Agropecuaria. 2020; 21(3):1-23. DOI:10.21930/rcta.vol21_num3_art:1552
CrossRef - Oliva-Hurtado M.M. Geospecial phytosanitary risk model: Xylella fastidiosa subsp. fastidiosa in Mexico. Thesis of master in Science, Colegio de Postgraduados, Estado de México, México, 2020.
CrossRef - Ibarra-Zapata E., Aguirre-Salado C. A., Miranda-Aragón L., Escoto-Rodríguez M., Loredo-Osh C., Mora Aguilera G., Casiano-Domínguez M., Aguirre-Salado A. I., Ramos-Méndez C., Villegas-Jiménez N., Urías-Morales C. R., González-Gómez R. Phytosanitary geospatial analysis of Musaceae fusariosis at a global level, with emphasis on Pantropical America. Investigaciones geográficas. 2021; 106: e60466. DOI: 10.14350/rig.60466.
CrossRef - Aguayo J., Cerf-Wendling I., Folscher A.B, Fourrier-Jeandel C., Ioos R., Mathews M. C., Mostert D., C. Renault, V. Wilson, and Viljoen A. First Report of Fusarium oxysporum f. sp. cubense Tropical Race 4 (TR4) causing Banana Wilt in the Island of Mayotte. Plant Dis. 2021;219. DOI: 10.1094/PDIS-06-20-1196-PDN.
CrossRef - Zakaria, L. Fusarium Species Associated with diseases of major tropical fruit crops. Horticulturae 2023; (9): 322. DOI:10.3390/horticulturae9030322
CrossRef - Gayol -Santana, L. Social dimension of the problem of the shortage of agricultural workers in the central zone of Puerto Rico: implications for the formulation of public policy. PhD. Thesis, University of Puerto Rico, San Juan, Puerto Rico, 2017.
- Amarie Magazine. https://amariemagazine.com/2019/08/13/secretario-de-agricultura-anuncia-medidas-para-atender-asunto-del-hongo-fusariym-oxysporum/(accessed on 29 july 2023).
- Zhou G.D., He P., Tian L., Xu S., Yang B., Liu L., Wang Y., Bai T., Li X., Li S. and Zheng S.J. Disentangling the resistant mechanism of Fusarium wilt TR4 interactions with different cultivars and its elicitor application. Front. Plant Sci. 2023; 14:1145837. DOI: 10.3389/fpls.2023.1145837
CrossRef

