Introduction
Pumpkins (Cucurbita pepo L., C. Maxima L. & C. Moshchata Duch.) are important vegetable crops in home-garden and commercial across the world. The pumpkin crop is one of the most popular summer vegetable crops grown on commercial scale worldwide. The crop is also widely grown in China, America, Mexico, and India. In terms of area and production, pumpkin is India’s second most important cultivated vegetable crop in the Cucurbitaceae family. In India, it is mostly grown in Madhya Pradesh, Uttar Pradesh, Chhattisgarh, West Bengal, Odisha, Karnataka, Tamil Nadu, Kerala, and Bihar. In India, the cultivated area is about 1.10 lakh ha, production is 23.12 million tonnes, and 21.01 tonnes/ha productivity during 2021-22 (agricoop.nic.in).
Begomoviruses (Geminiviridae family) are circular single-stranded DNA viruses with twinned (geminate) morphology and contain either monopartite (DNA-A) or bipartite (DNA-A and DNA-B) genomes approx. 2.6kb to 2.8kb in size. Begomovirus is transmitted by vector Bemisia tabaci (Genn.), an emerging plant virus pathogen in the family Geminiviridae, which inflicts diseases on a variety of crops in tropical and subtropical areas.1 Pumpkin is a common host of various begomoviruses.2,3,4,5,6,7 In northern and southern India, mosaic and leaf curl diseases caused by begomoviruses, are rising problems because they affect the production and yield of pumpkin crops. The current study aims to determine the prevalence and distribution of begomovirus infections in Central India especially in Madhya Pradesh state. Symptomatic mosaic leaf samples were collected for the detection and identification of associated begomovirus on pumpkin crops. The data collected will be crucial for the improvement of virus-resistant varieties/cultivars and effective management plans for begomovirus infections affecting pumpkin crop species in Central India.
Materials and methods
Investigation for disease incidence
The survey was conducted from different locations in the Bhopal district. In each location, symptomatic and healthy plant leaves were collected.
Genomic extraction and detection of Begomovirus using polymerase chain reaction
The method described by Dellaporta et al. (1983) was used to extract the total DNA from leaf samples taken from both symptomatic and healthy plants. To amplify the coat-protein gene of begomovirus, the polymerase chain reaction (PCR) was carried out using total DNA as a template and a set of begomovirus degenerate CP region primers: CPIT-I/ CPIT-T.8 The PCR was performed in 25 μl volume, containing template DNA (100 ng), 10X Taq buffer with 25 mM MgCl2, 10 mM dNTPs, 25 pmol primers (each), and Taq DNA polymerase (250 U, DSS Takara Bio India Pvt. Ltd., New Delhi). The reactions were performed in a BIO-RAD T-100 Thermal cycler at 94°C for 5 min followed by 35 cycles of 94°C for 1 min, 47°C for 1 min, and 72°C for 1:30 min. The cycling was succeeded by a final extension for 5 min at 72°C. The PCR amplicons were analysed by 1.2% agarose gel electrophoresis and the expected size was eluted using Wizard SV Gel and PCR Clean-up System Promega, USA.
Sequencing and Sequence Analysis
The eluted product was sequenced commercially by Barcode Biosciences Pvt. Ltd, Bengaluru (India) using both the forward and the reverse primers. BLAST (Basic Local Alignment Search Tool) search analysis of nucleotide sequence of the virus isolates was conducted using Blastn with sequences available in the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE). The Genomatix alignment tool and the Sequence Demarcation Tool (SDTv1.2) were used for the pairwise identity comparison of virus isolates under study with the sequences of selected begomovirus obtained from the GenBank database.9,10 Sequences under study were aligned using the ClastalW algorithm and MEGAv11; bootstrapping for 1000 replicates was used for generating phylogenetic trees.11,12
Result
Disease symptoms and effectiveness of transmission
During the year 2022, in five fields in Bhopal District, Madhya Pradesh, India (two fields at Berkhedi bazyaft (23.177̊ N, 77.340̊ E), and one-one sample collected field at Shyampur (23.341̊ N, 77.426̊ E), Arwaliya (23.343̊ N, 77.412̊ E), and Saket Nagar (23.208̊ N, 77.456̊ E)), virus-like symptoms were observed in pumpkin crops. In each field visited infected leaf samples showing Severe Mosaic, Mosaic, and leaf curling were collected (Figure 1).
 |
Figure 1: Healthy and Symptomatic leaves of pumpkin: (A) Healthy leaf (B) Leaf curling (C) Severe Mosaic (D) Mild Mosaic (E) Leaf curling with severe mosaic (F) Mosaic. |
Genomic extraction and detection of Begomovirus using PCR
Total genomic DNA was isolated by the Dellaporta method from symptomatic and asymptomatic leaf samples to confirm the association of begomovirus with leaf curl and mosaic disease of pumpkin crops.13 This method yielded a good quality & quantity of DNA preparation as checked by taking its best O.D. at 260/280 is 1.8 and concentration also checked by the 1.2% agarose gel electrophoresis. DNA samples were subjected to PCR using the begomovirus coat-protein gene-specific primers for DNA-A: CPIT-I/CPIT-T8 primers amplified coat protein (AV1) gene. The PCR products were analyzed by electrophoresis in 1.2% agarose gel. As expected, bands of ~800 bp (CPIT-I/CPIT-T). The PCR amplicon of ~800 bp of the PCR reaction was gel eluted through Wizard SV Gel and PCR Clean-Up System (Promega Pvt. Ltd. USA), and eluted products sequenced from Barcode Biosciences Pvt. Ltd, India. The obtained sequence data of the complete coat protein gene (AV1) were deposited in the NCBI GenBank database under accession numbers OQ320768, OQ320770, and OQ116978 in C. maxima, OQ320774 in C. moshchata, and OQ116977 C. pepo.
Sequencing and Sequence Analysis
BLASTn analysis of the understudy pumpkin virus isolates coat protein gene (Acc. No. OQ320768, OQ320770, OQ320774, and OQ116977, OQ116978) showed 97.67-99.74% identities with each other and showed the highest 95-97% sequence identity with isolates of Tomato leaf curl New Delhi virus (ToLCNDV: MH577015, KF551576) in tomato and 70-95% sequence identity with Squash leaf curl China virus (SLCCNV: AY396151, LC511776, MW248689); Pumpkin yellow vein mosaic virus (PYVMV: AY686500); Tomato leaf curl Palampur virus (ToLCPalV: FJ931537, GQ225738); Squash leaf curl Philippines virus (SLCPHV: AB085793, DQ866135); Squash leaf curl virus (SLCV: KY652743); and Chilli leaf curl virus (ChiLCV: MN119490).
Pairwise sequence comparisons (By Genomatix DiAlign Tool) showed the highest 95% similarities to the ToLCNDV and 70-72% ChiLCV (Table 1). The AV1 (Coat-protein) gene of begomovirus isolates was compared with other begomoviruses available in the GenBank database, and pairwise identity scores were calculated using the SDTv1.2. The SDT analysis showed that isolates OQ320768, OQ320770, and OQ320774 had maximum nucleotide (nt) similarities (95%); OQ116977 had maximum nt similarities (93%), and OQ116978 had maximum similarities (94%) with ToLCNDV. The isolates individual coat-protein gene (AV1) was compared with the AV1 of different begomovirus. The analysis showed that AV1 shared the highest 80-99% amino-acid (aa) similarities with ToLCNDV from India. This isolates species was also supported by a two-dimensional color-coded matrix of pairwise identity scores of the AV1 gene of ToLCNDV generated by SDTv1.2 (Figure 2).
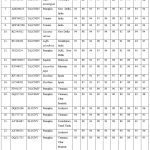 |
Table 1: The Genomatix DiAlign programme was used to check similarity at nucleotide (nt) and amino acid (aa) of coat-protein begomovirus isolates under study (OQ116977, OQ116978, OQ320768, OQ320770, and OQ320774) with selected begomoviruses. |
 |
Figure 2: Using the Sequence Demarcation Tool, a two-dimensional color-coded matrix of pairwise identity scores of begomovirus isolates was constructed. (http://web.cbio.uct.ac.za/SDT). |
To determine the evolutionary relationship of the understudy begomovirus isolates (OQ320768, OQ320770, OQ320774, OQ116977, and OQ116978), the phylogenetic trees were constructed using the MEGAv11 using the neighbor-joining method with 1000 bootstraps replicates with selected begomoviruses that were found to be closely related by BLASTn (Figure 3). The understudy begomovirus isolates (OQ320768, OQ320770, OQ320774, OQ116977, and OQ116978) showed the closest relationship with the ToLCNDV (MH577015) on tomato plant from Anand, Gujarat, India and shared distinct relationships with other begomovirus species reported in tomato, pumpkin, Zucchini and other plant species from India and abroad.
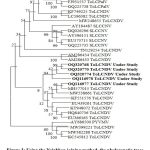 |
Figure 3: Using the Neighbor-joining method, the phylogenetic trees constructed from aligned AV1 nucleotide sequences of pumpkin isolates with other begomoviruses. |
Discussion
During the summer, rainy, and winter seasons in various areas of India, pumpkin is one of the most extensively grown cucurbitaceous vegetables. The pumpkin crop is affected by many insects and diseases caused by fungi, bacteria, and viruses. Globally, agriculture is severely impacted by begomoviruses spread by whiteflies because of their variety, capacity for recombination, fluctuating host ranges, and severe disease symptoms.14 In the current investigation, leaf samples of C. pepo, C. maxima, and C. moshchata that had mild to severe mosaic and leaf curling were collected from the field and examined by PCR for the presence of the begomovirus using a primer that was specific to the coat-protein gene of the begomovirus. Further begomovirus-positive pumpkin samples were sequenced and submitted in Genebank database. The complete coat protein gene sequences of understudy virus isolate showed the highest similarity with isolates of ToLCNDV and infecting tomato (MH577015; KF551576) and ridge gourd (MW538661) from India.
An emerging problem of ToLCNDV affects a number of crops in India, Pakistan, the Philippines, Thailand, Italy, and China.15,16,29 However, ToLCNDV is the most important viral pathogen in the solanaceae family, it has been observed in northern, north-western and southern India infecting a variety of cucurbitaceous crops, including pumpkin, ridge gourd, bottle gourd, long melon, watermelon, ivy gourd, bitter gourd, and cucumber.
In the early 1940s, a yellow vein disease infestation was first identified in crops from northern India.2 The causal virus is designated as “Pumpkin Yellow Vein Mosaic Disease” (PYVMD) and the occurrence of this disease was first reported from New Delhi,23,15 Pune,3 Maharashtra,3 Uttar Pradesh,24, 27 West Bengal,25 Kerala,26 Karnataka,4, 6 Coimbatore,5 Assam,7 and Bihar.28
On the basis of the highest sequence identities, pairwise similarities, and closest phylogenetic relationship understudy virus isolate was identified as an isolate of Tomato leaf curl New Delhi virus on the pumpkin. To our knowledge, this was the first recode of Tomato leaf curl New Delhi virus associated with mosaic disuse of pumpkin from Central India.
Pumpkin plant species are perennial climber crops grown all over India, especially in the Northern and Central parts. The molecular data presented here provide useful information on the occurrence of ToLCNDV in pumpkin plant species from Central India. Therefore further studies are required to know the role of climate change in favor of the persistence of vector and spread of begomovirus and the development of infectious clones to screen the begomovirus disease on pumpkins. Further, more molecular studies are required in the future for the characterization of associated begomoviruses on the basis of their complete genome and to develop management strategies to control the mosaic disease of pumpkins to improve their quality and production of pumpkin crops in India.
Acknowledgment
The authors are thankful to the Vice Chancellor and Head (Department of Microbiology), Barkatullah University, Bhopal, India, for the use of the facilities.
Conflict of Interest
There is no conflict of interest.
Funding Source
The present study receives no funding.
Reference
- Brown JK. The molecular epidemiology of begomoviruses. In: Khan JA and Dykstra J. Trends in plant virology. The Haworth Press, New York. 2001; 279-316.
CrossRef - Vasudeva RS, and Lal TB. A mosaic disease of bottle gourd. Indian J Agric Sci. 1943;13:182-191.
- Capoor SP, and Ahmad RU. Yellow vein mosaic disease of field pumpkin and its relationship with the vector, Bemisia tabaci. Indian Phytopath. 1975;28:241-246.
- Jayashree K, Pun KB, Doraiswamy S. Virus-vector relationship of yellow vein mosaic virus and whitefly (Bemisia tabaci) in pumpkin. Indian Phytopath. 1999;52:10-13.
- Maruthi MN, Rekha AR, and Muniyappa V. Pumpkin yellow vein mosaic disease is caused by two distinct begomoviruses: complete viral sequences and comparative transmission by an indigenous Bemisia tabaci and the introduced B‐biotype. EPPO bulletin. 2007;37(2):412-419. https://doi.org/10.1111/j.1365-2338.2007.01127.x
CrossRef - Muniyappa V, Maruthi MN, Babitha CR, Colvin J, Briddon RW, Rangaswamy KT. Characterization of pumpkin yellow vein mosaic virus from India. Ann. Appl. Biol., 2003;142:323-331. https://doi.org/10.1111/j.1744-7348.2003.tb00257.x
CrossRef - Baldodiya GM, Devi K, Borah BK, Nath PD, Modi Characterization and in silico proteomic analysis of C2 and C3 proteins of squash leaf curl China virus associated with pumpkin leaf curl disease in Assam, India. Acta Virol., 2019;63(2):139-148. DOI: 10.4149/av_2019_202
CrossRef - Singh R. Molecular characterization of a virus yellow mosaic disease in Cucurbita maxima and development of diagnostics for detection of virus. Diss. PhD thesis, Lucknow University, Lucknow, India, 2005.
- Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics (Oxford, England), 1999; 15(3):211-218.
CrossRef - Muhire BM, Varsani A, Martin DP, Kuhn JH. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS One. 2014;9(9):e108277.
CrossRef - Thompson JD, Higgins DG, and Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673-4680. DOI: 1093/nar/22.22.4673
CrossRef - Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol., 2007; 24:1596-1599.
CrossRef - Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1,19-21. DOI: 1093/molbev/msm092
CrossRef - Varma A, and Malathi VG. Emerging geminivirus problems: a serious threat to crop production. Ann Appl Biol. 2003;142:145-164. https://doi.org/10.1111/j.1744-7348.2003.tb00240.x
CrossRef - Phaneendra C, Rao KS, Jain RK, Mandal B. Tomato leaf curl New Delhi virusis Associated With Pumpkin Leaf Curl: A New Disease in Northern India. Virus Disease. 2012;23(1):42-45. Doi: 10.1007/s13337-011-0054-z
CrossRef - Parrella G, Troiano E, Formisano G, Accotto GP, Giorgini M. First report of Tomato leaf curl New Delhi virus associated with severe mosaic of pumpkin in Italy. Plant Dis. 2018;102(2):459. https://doi.org/10.1094/PDIS-07-17-0940-PDN
CrossRef - Mandal B, Mandal S, Sohrab SS, Pun KB, Varma A. A new yellow mosaic disease of chayote in India. Plant Pathol., 2004;53:797. https://doi.org/10.1111/j.1365-3059.2004.01075.x
CrossRef - Khan MS, Raj SK, and Singh R. First report of tomato leaf curl New Delhi virus infecteing chilli in India. Plant Pathol. 2006;55:289. https://doi.org/10.1111/j.1365-3059.2006.01324.x
CrossRef - Sohrab SS, Mandal B, Ali A, Varma A. Molecular diagnosis of emerging begomovirus diseases in cucurbits occurring in northern India. Indian J Virol. 2006;17(2):88-95.
- Tiwari AK, Sharma PK, Khan MS, Snehi SK, Raj SK, Rao GP. Molecular detection and identification of Tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in Bitter gourd (Momordica charantia), a medicinally important plant in India. Medicinal Plants. 2010;2(2):117-123. Doi : 10.5958/j.0975-4261.2.2.018
CrossRef - Raj SK, Snehi SK, Khan MS, Tiwari AK, Rao GP. Molecular identification of Ageratum enation virus associated with mosaic disease of pointed gourd (Trichosanthes dioica ) in India. Phytoparasitica. 2011;39:497-502. DOI 10.1007/s12600-011-0182-4
CrossRef - Nagendran K, Mohankumar S, Aravintharaj R, Balaji CG, Manoranjitham SK, Singh AK, Rai AB, Singh B, Karthikeyan G. The occurrence and distribution of major viruses infecting cucurbits in Tamil Nadu state, India. Crop Prot. 2017;99:10-16. https://doi.org/10.1016/j.cropro.2017.05.006
CrossRef - Varma PM. Ability of the whitefly to carry more than one virus simultaneously. Curr. Sci. 1955;24:317-318.
- Bhargava B, and Bhargava KS. Cucurbit mosaic viruses in Gorakpur. Indian J Agric Sci. 1977;47:1-5.
- Ghosh SK, and Mukhopadhyay S. Viruses of pumpkin (Cucurbita moschata) in West Bengal. J. Phytopathol. 1979;94:172-184. https://doi.org/10.1111/j.1439-0434.1979.tb01547.x
CrossRef - Latha P, and Gopalakrishnan TR. Inheritance of mosaic disease and silvery leaf trait in pumpkin (Cucurbita moschata Poir.). Agric Res J Kerala, 1993;6:97-101. DOI:17660/ActaHortic.2000.510.48
CrossRef - Tiwari AK, Snehi SK, Singh R, Raj SK, Rao GP, Sharma PK. Molecular identification and genetic diversity among six Begomovirus isolates affecting cultivation of cucurbitaceous crops in Uttar Pradesh, India. Phytopathol. Pflanzenschutz. 2011;45(1):62-72. https://doi.org/10.1080/03235400903458803
CrossRef - Kumar R, Srinivasaraghavan A, and Bharti J. Survey of Begomovirus affecting cucurbits in Bihar. The Pharma Innovation Journal. 2021;10(7):1735-1743.
- Gu Q, Yan L, Liu L, Bao W, Fang H, Xu J, Li J, Kang B, Wu H, Wang K, Tao X, Peng B. First report of tomato leaf curl New Delhi virus infecting several cucurbit plants in China. Plant Dis. 2023. doi: 10.1094/PDIS-01-23-0180-PDN
CrossRef

