Introduction
The scented rice of Manipur, Chakhao, meaning delicious rice in Manipuri language, possess high socio-cultural values and is in close relationship with spiritual cultural practices of Meiteis. Chakhao has a unique flavour and aroma. It is served as a special item in cultural and religious ceremonies such as Usob (death ceremonies), Kang-pali (religious festival), Chak-umba (first rice-eating ceremony)1. Chakhao, an indigenous rice variety of Manipur, is nutrient-rich and contains essential amino acids, minerals, and carotenoids. It may help prevent cancer, diabetes, heart disease, and Alzheimer’s disease2-3. Chakhao is grown at certain parts of Imphal valley in Manipur. Due to its poor yield, Chakhao is grown in very limited acreage by farmers in Manipur for ceremonial and cultural purposes. There is a pressing need to increase its output because it is becoming more and more popular due to its nutraceutical characteristics and export demand4.
In modern agriculture, synthetic agrochemicals are used for enhancing crop production. The dependence on agrochemicals for improving crop production pose serious risks on ecological and human health5. Rhizobacteria are associated with the soils near the roots and intimately interact with the exudates of the roots6. Plant growth promoting rhizobacteria (PGPR) are among the beneficial soil microorganisms found in the rhizospheric region of the plant7. Intensive research on plant growth promoting bacteria (PGPB) is underway worldwide for developing biofertilizers and biocontrol agents (BCAs) as better alternatives to agrochemicals, as the latter harm the environment and human health besides possess the burden of high costs to poor farmers8. Biological control is slow but can be long lasting, inexpensive, and harmless to living organisms and the ecosystem; it neither eliminates the pathogen nor the disease, but brings them into natural balance9. As most PGPBs show inconsistent performance in the field conditions, there is urgent need for survey of indigenous strains suited to local conditions. The use of bacteria having biocontrol and PGP activities is urgently warranted to increase Chakhao growth10.
Certain bacterial taxa e.g., actinomycetes and Bacillus spp have been shown to play a significant function in the plant rhizosphere by secreting a variety of antimicrobial substances that inhibit the growth of common root diseases. It is reported that rhizospheric bacteria and other beneficial bacteria have the ability to solubilize phosphate and can produce indole acetic acid (IAA), siderophore, ammonia, 1- aminocyclopropane-1-carboxylic acid (ACC) deaminase and cell wall degrading enzymes such as chitinase, glucanase and protease11-16. Rhizospheric isolates associated with Chakhao varieties may have potential to produce unique metabolites, which can be exploited for application in agricultural, pharmaceutical and other industries.
The present study aims at investigating the antifungal and biofertilizing potential of the rhizospheric isolates from 6 different Chakhao cultivars found in Manipur17.
Materials and Methods
Soil Sampling and Bacterial Isolation
Rhizospheric bacteria were isolated from all 6 rhizospheric soil samples collected from Wairi, Imphal East (93.88E, 24.78N) and Lilong Leihaokhong, Thoubal (93.93E, 21.70N), using 3 different media i.e., Starch Casein Nitrate Agar (SCNA), Nutrient Agar (NA) and Gauze Medium No. 1 (GM1).
1 g of the soil sample was added to 99 ml of sterile distilled water (SDW). The sample was thoroughly vortexed (150 rpm, 30°C, 10 min). Serial dilutions of the soil suspensions were then performed (10-2 to 10-6). The aliquots of various dilutions from 10-2 to 10-6 were spread plated on agar plates. The isolation plates were incubated for 3-4 days at 30°C. Morphologically distinct bacterial colonies were sub-cultured till pure cultures were obtained18.
Antifungal Assay
Antifungal assay of the rhizospheric bacteria were carried out by dual culture method19-20. The inhibition zones are due to diffusible compounds by the bacterial isolate in the antifungal assay. After the full growth of fungal mycelia on the control plates, the inhibition in the remaining plates was measured. The percentage of mycelial growth inhibitions was calculated using the formula:
![]()
Where,
R represents the radial growth of the test pathogen in the control plates (measured in mm), and r is the radial growth of the test pathogen in the test plates.
Screening of Plant Growth Promoting (PGP) Traits
Indole Acetic Acid (IAA) Production Test
IAA production was determined by inoculating bacterial isolates in NB medium containing 2 mg/ml of L-tryptophan, (150 rpm, 30ºC, 5 d)21. The fully grown culture broth was centrifuged (10,000 rpm, 10 min) and 1 ml of the supernatant was mixed with 2 ml of Salkowski’s reagent. A pink colour indicates a positive test for IAA production.
For the quantitative assay, the bacterial isolates were inoculated in NB containing 2 mg/mL of Tryptophan under shaking conditions (150 rpm, 30ºC). 5 mL aliquot was withdrawn periodically from each culture flask at 24 hours intervals and centrifuged at 10,000 rpm for 10 min21. 1 mL of the supernatant was mixed with 2 mL of Salkowski’s reagent and incubated at 30ºC for 20 min. The absorbance was measured at 530 nm and the amount of IAA produced was calculated by comparing it with the standard IAA curve.
Phosphate Solubilization Test
Spot inoculation on NBRIP-BP (National Botanical Research Institute’s Phosphate Growth Medium) was carried out to determine phosphate solubilization by bacterial isolates. A halo zone around the bacterial colony after 4 days of incubation at 30oC indicated a positive test22.
Quantitative estimation of phosphate solubilization was done by inoculating in 100 mL of NBRIP-BP medium (pH 7). It was then incubated in a shaker incubator (150 rpm, 30°C). 5 mL aliquot withdrawn periodically at 24 hours intervals was centrifuged (10,000 rpm, 10 min) and the supernatant was analysed for pH and Phosphate concentration23. KH2PO4 was used as the standard24.
Siderophore Production Test
SCNA (without iron) amended with CAS-substrate (Chrome Azurol S) were inoculated with 6 mm agar plugs of bacterial isolate and incubated at 30°C for 7 days. The formation of an orange colour halo surrounding the colony was considered a positive result for siderophore production25.
Quantitative estimation of siderophore production was done on five different iron deficient liquid media: Starch casein nutrient broth (SCNB), Casamino acid medium (CAA), Nutrient broth (NB), Succinic acid medium (SM) and Bharbhiaya and Rao medium (BR) by CAS-shuttle assay26-27. 5 mL aliquots were withdrawn periodically at 24 hours intervals and centrifuged (10,000 rpm, 10 min). An equal volume of CAS reagent was added to the supernatant. Absorbance at 630 nm was noted. A reference made of 1 mL uninoculated broth and 1 mL CAS reagent was used. The amount of siderophore produced (percentage siderophore units) was calculated by using the formula:
![]()
Where,
Ar represents the absorbance of reference and As represents the absorbance of the sample at 630 nm.
Ammonia Production Test
The bacterial isolates were inoculated in 10ml of peptone water and incubated in a shaker incubator (150 rpm, 30°C) for 4 days. To each test tube, 0.5 ml of Nessler reagent was then added. A colour change of brown to yellow indicated ammonia production by the bacterial isolates28.
1-Aminocyclopropane-1-Carboxylic Acid (ACC) Deaminase Production Test
Nitrogen-free Dworkin and Foster’s (1958), DF salts minimal agar medium supplemented with 2 g of (NH4)2SO4 as a sole nitrogen source was used for screening ACC deaminase production29-30. Isolates were inoculated on the media and then incubated at 30°C for 4 days. Bacterial growth in the plates indicated a positive test for ACC deaminase production.
In Vitro Seed Germination (Vigor Index)
In vitro rice seed germination by the isolates were carried out according to Tamreihao et al. (2016)31. The bacterial isolates were inoculated in NB and incubated for 3 days to get fully grown cultures. It was then centrifuged (10,000 rpm, 10 min). The pellets were collected and washed thrice with SDW (Sterile Distilled Water) and culture suspensions were prepared using SDW. Black rice (Variety: Chakhao Amubi) seeds were surface sterilized with 70% ethanol for 5 min, followed by 0.2% sodium hypochlorite for 5 min, and rinsed four times with SDW. Surface sterilized seeds were soaked in the cell suspensions and kept under shaking conditions (150 rpm, 30°C) for 2 hours. The seeds were then transferred to sterile plates containing wetted filter papers (10 seeds per plate). Untreated sterile seeds soaked in SDW were used as a control. The plates were then incubated at 28°C for 5 days. The number of germinated seeds, roots and shoot lengths were measured and vigor indices were calculated using the following formula32;
![]()
Where,
Seedling length= shoot length + root length
Statistical Analysis
All the data were subjected to one-way analysis of variance (ANOVA) followed by independent t-test (p≤0.05) using the SPSS software.
Strain Identification
Genomic DNA extraction and PCR amplification of 16S rDNA sequences were performed33. The PCR products obtained were sent to AgriGenome Labs Pvt. Ltd. (Kerala, India) for sequencing. The bacterial sequences were subjected to BLAST alignment analysis using the NCBI GenBank database to obtain the accession number34.
Results
Isolation
Altogether, 323 morphologically distinct isolates were obtained from rhizospheric soils of six (6) different cultivars of Chakhao. These isolates were preserved in 30% glycerol stock at − 20°C for further studies.
Antifungal Assay
All the 323 isolates were screened for antifungal assays against the test fungal pathogens, of which 64 showed antifungal activities against one or more fungal pathogens. Four (4) isolates (CR12, CW11, CA2 and CP2) exhibited potent antifungal activity (Figs. 1-2 and Table 1).
 |
Figure 1: Antifungal activities of the 4 best strains against the fungal pathogens (a-y) |
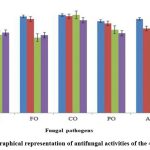 |
Figure 2: Graphical representation of antifungal activities of the 4 best strains |
Table 1: Antifungal assay of the rhizospheric bacterial isolates
| Sl. No. | Bacterial Isolates | Percentage Inhibition Zone | ||||
| MTCC4633 (RS) | MTCC287 (FO) | MTCC2605 (CO) | MTCC1477 (PO) | MTCC1344 (AN) | ||
| 1. | CA1 | 67.4 | – | 69.3 | – | – |
| 2. | CA2 | 73.75 | 71.25 | 92.5 | 78.5 | 78.75 |
| 3. | CA3 | – | 65.3 | 57.2 | – | – |
| 4. | CA6 | 45.4 | – | 51.6 | – | – |
| 5. | CA8 | – | 60.4 | 71.6 | – | – |
| 6. | CA11 | 52 | – | – | – | – |
| 7. | CA12 | – | 65 | – | – | – |
| 8. | CA14 | – | 43.7 | 61.4 | – | – |
| 9. | CA21 | 47.3 | – | – | – | – |
| 10. | CA23 | – | 56.8 | – | – | – |
| 11. | CA28 | – | – | 71 | – | 67.3 |
| 12. | CA33 | 64.3 | – | – | – | – |
| 13. | CA37 | – | – | 53.2 | – | – |
| 14. | CA45 | 47.2 | – | – | 34.3 | – |
| 15. | CA46 | – | 61 | – | – | – |
| 16. | CA51 | – | – | 56.4 | – | – |
| 17. | CA54 | 53.2 | – | 55.6 | – | – |
| 18. | CR1 | 43 | – | 51.5 | – | – |
| 19. | CR3 | – | 54 | 61.4 | – | – |
| 20. | CR8 | 37 | 48.5 | – | – | – |
| 21. | CR10 | 45 | 53.3 | – | – | – |
| 22. | CR12 | 93.4 | 91.2 | 92.5 | 86.7 | 88.8 |
| 23. | CR13 | 56.3 | – | 62.3 | – | – |
| 24. | CR19 | – | 57.4 | – | – | – |
| 25. | CR25 | – | – | 80.6 | – | – |
| 26. | CR26 | – | – | – | 45.3 | 54.2 |
| 27. | CR31 | 58.3 | – | 60.3 | – | – |
| 28. | CR35 | 34 | – | 42 | – | – |
| 29. | CR41 | – | – | 56.8 | – | – |
| 30. | CR42 | – | 57.3 | – | – | – |
| 31. | CR53 | 46.7 | – | – | – | – |
| 32. | CR58 | – | 35 | 46.3 | – | – |
| 33. | CR60 | – | – | – | – | 65.4 |
| 34. | CR62 | – | – | – | 43.6 | – |
| 35. | CR65 | – | – | 79.5 | – | – |
| 36. | CR68 | – | 45.6 | 57.5 | – | – |
| 37. | CR71 | 76.3 | – | – | – | – |
| 38. | CR74 | – | – | 65.4 | – | – |
| 39. | CP2 | 75.8 | 73.4 | 87.6 | 75.3 | 77.3 |
| 40. | CP4 | – | 54.8 | 67.4 | – | – |
| 41. | CP12 | 46 | – | – | – | – |
| 42. | CP16 | – | – | – | 56.4 | 67 |
| 43. | CP22 | 43 | 54.5 | – | – | – |
| 44. | CP29 | 41.4 | 46.7 | – | – | – |
| 45. | CP31 | – | – | 73.7 | – | – |
| 46. | CW7 | – | – | – | 65.4 | 43.6 |
| 47. | CW8 | – | – | 64.8 | – | – |
| 48. | CW11 | 90.4 | 88.7 | 91.3 | 84.5 | 79.7 |
| 49. | CW13 | 84.3 | 78.6 | – | – | – |
| 50. | CW24 | – | 49 | 58.4 | – | – |
| 51. | CW25 | 72 | – | – | – | – |
| 52. | CW34 | – | – | 69.6 | – | – |
| 53. | CW41 | – | – | – | – | 77 |
| 54. | CS5 | – | – | 37.6 | – | – |
| 55. | CS9 | 53.2 | – | 47.4 | – | – |
| 56. | CS13 | 81.4 | 75.8 | 80.6 | – | – |
| 57. | CS23 | 72 | – | – | – | – |
| 58. | CS32 | – | 43.6 | 35 | – | – |
| 59. | CS40 | – | – | 56.9 | – | – |
| 60. | CAng4 | 74 | – | 57.8 | – | – |
| 61. | CAng11 | – | – | – | 43 | 37.8 |
| 62. | CAng16 | 29 | – | – | – | – |
| 63. | CAng24 | – | – | 43.4 | – | – |
| 64. | CAng31 | – | – | 46.7 | – | – |
Plant growth promoting (PGP) activities
All the isolates were then subjected to screening for PGP traits such as IAA, siderophore, ammonia and ACC deaminase production, and phosphate solubilization. 69 isolates showed positive results for one or more PGP traits (Table 2).
Table 2: PGP assays of rhizospheric bacterial isolates
| Sl. No. | Bacterial Isolates | Plant growth promoting traits | ||||
| IAA production | Phosphate solubilization | Siderophore production | Ammonia production | ACC production | ||
| 1. | CA1 | + | – | – | + | + |
| 2. | CA2 | + | + | + | + | + |
| 3. | CA3 | – | – | – | + | – |
| 4. | CA7 | + | – | – | – | + |
| 5. | CA8 | – | – | – | + | – |
| 6. | CA10 | + | – | – | + | – |
| 7. | CA12 | – | – | – | + | – |
| 8. | CA14 | + | – | – | – | – |
| 9. | CA21 | – | + | – | – | + |
| 10. | CA22 | – | – | – | + | – |
| 11. | CA25 | + | – | – | – | – |
| 12. | CA28 | – | + | – | – | + |
| 13. | CA31 | + | – | – | + | – |
| 14. | CA37 | – | – | – | – | + |
| 15. | CA39 | + | – | – | – | – |
| 16. | CA41 | – | – | – | + | – |
| 17. | CA44 | – | – | – | – | + |
| 18. | CA53 | – | – | – | – | + |
| 19. | CA54 | – | – | – | – | + |
| 20. | CA57 | – | – | – | + | – |
| 21. | CA60 | – | – | + | – | – |
| 22. | CA62 | – | – | – | + | + |
| 23. | CA69 | + | – | – | + | – |
| 24. | CR1 | + | – | – | + | – |
| 25. | CR2 | + | – | – | + | – |
| 26. | CR4 | – | + | – | – | – |
| 27. | CR7 | – | + | + | – | – |
| 28. | CR8 | – | – | – | + | – |
| 29. | CR10 | – | – | – | + | + |
| 30. | CR12 | + | + | + | + | + |
| 31. | CR21 | – | + | – | – | – |
| 32. | CR26 | – | – | – | + | – |
| 33. | CR32 | – | – | – | + | – |
| 34. | CR37 | + | – | – | – | + |
| 35. | CR39 | + | – | – | – | + |
| 36. | CR45 | – | + | – | – | + |
| 37. | CR47 | – | – | + | + | – |
| 38. | CR54 | – | – | – | + | – |
| 39. | CR63 | – | – | – | – | + |
| 40. | CR68 | – | – | – | – | + |
| 41. | CR71 | – | – | – | + | – |
| 42. | CR75 | – | – | – | + | – |
| 43. | CR77 | – | – | – | + | + |
| 44. | CP2 | + | + | + | + | + |
| 45. | CP3 | + | – | – | + | – |
| 46. | CP7 | + | – | – | – | – |
| 47. | CP8 | – | + | – | + | – |
| 48. | CP13 | – | – | – | + | – |
| 49. | CP16 | – | – | – | – | + |
| 50. | CP24 | – | – | – | + | + |
| 51. | CW1 | – | – | – | + | – |
| 52. | CW2 | – | – | – | + | – |
| 53. | CW4 | + | – | – | – | + |
| 54. | CW6 | – | – | – | – | + |
| 55. | CW11 | + | + | + | + | + |
| 56. | CW13 | – | + | – | + | – |
| 57. | CW27 | – | – | – | + | + |
| 58. | CW45 | + | + | – | – | – |
| 59. | CS4 | – | – | – | + | – |
| 60. | CS6 | – | – | – | + | – |
| 61. | CS13 | + | – | + | – | + |
| 62. | CS28 | + | – | – | – | + |
| 63. | CS37 | + | – | – | – | + |
| 64. | CS42 | – | + | + | – | – |
| 65. | CAng1 | + | – | – | – | – |
| 66. | CAng4 | + | – | – | – | – |
| 67. | CAng9 | – | + | – | – | – |
| 68. | CAng24 | – | – | – | + | – |
| 69. | CAng31 | – | – | – | + | – |
Shortlisting of promising isolates
Based on the screening of antifungal and PGP activities, 4 most potent isolates (CR12, CW11, CA2 and CP2) were shortlisted for molecular characterization and seedling vigor studies.
Quantitative estimation of PGP activities
The shortlisted potent strains were subjected to quantitative estimation of PGP activities like phosphate solubilization, IAA and siderophore production. The strain CR12 produced the highest amount of IAA (73 μg/mL) followed by CW11 (71.4 μg/mL), CP2 (70.5 μg/mL) and CA2 (62.91 μg/mL) respectively after 10 d of incubation. The strain CA2 could solubilize the highest amount of inorganic P ( up to 200 μg/mL) after 3 d of incubation with concomitant decrease in medium pH (from 5.5 to 2.69) followed by CR12, CW11 and CP2 ( up to 156.006, 113.5 and 105.7 μg/mL respectively). CA2 showed maximum siderophore production (74.37% siderophore units in SCNB) followed by CR12, CP2 and CW11 (73.15% in NB, 67.80% in SCNB and 65.50% in NB respectively) after 6 d of incubation.
Characterization of promising strains
Strain CR12 showed the highest 16S sequence similarity with Bacillus subtilis, CW11 with Bacillus paralicheniformis, CA2 with Bacillus sp. CCMB1014 and CP2 with Bacillus licheniformis. They were deposited in NCBI to obtain the accession numbers. Hence, they are designated as Bacillus subtilis strain CR12 (Accn. No.OM866257), Bacillus paralicheniformis strain CW11 (Accn. No.OM868047), Bacillus sp. CCMB1014 strain CA2 (Accn. No.OM868070) and Bacillus licheniformis strain CP2 (Accn. No. OM892495).
In Vitro Seed Germination (Vigor Index)
Seedling vigor assays for the 4 selected promising strains (CR12, CW11, CA2, and CP2) were done on Chakhao Amubi. The isolates exhibited higher vigor indices (CR12: 668.00, CW11: 478.91, CA2: 618.66 and CP2: 510.92) relative to the control (164.48) as shown in Fig. 3 and Table 3.
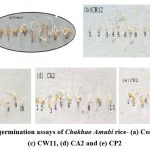 |
Figure 3: Seed germination assays of Chakhao Amubi rice- (a) Control, (b) CR12, (c) CW11, (d) CA2 and (e) CP2 |
Table 3: Vigor indices of the shortlisted best strains against control
| Treatments | Fresh weight (gm) | Dry weight (gm) | Root length (cm) | Shoot length (cm) | % seed germination | Vigor index |
| Control | 0.07±0.01b | 0.02±0.00b | 1.42±0.78b | 0.95±0.53b | 69.4±2b | 164.48 |
| CR12 | 0.09±0.01a | 0.03±6.01a | 4.29±0.7a | 3.39±0.53a | 86.98±1.2a | 668.00 |
| CW11 | 0.06±0.00c | 0.02±0.00b | 3.7±0.74c | 2.70±0.48c | 74.83±1.88c | 478.91 |
| CA2 | 0.07±0.00b | 0.03±0.00a | 4.29±0.77a | 3.38±0.53a | 80.66±9.90d | 618.66 |
| CP2 | 0.06±0.00c | 0.02±0.00b | 3.83±0.67c | 3.04±0.47a | 74.37±2.13c | 510.92 |
*Values with different alphabet within a column are significant at P≤0.05
Discussion
According to a survey of Chakhao rice cultivation in some selected villages of Manipur by Borah et al. (2018), it was found that the yield of Chakhao is generally lower (~1.3-1.8 tons/hectare) as compared to hybrid and traditional rice varieties (~2.0-5.5 tons/hectare)35. Farmers generally do not apply chemical fertilizers and farmyard manure in the cultivation of Chakhao rice as application of excess inorganic and organic fertilizer increases lodging and sterility1. So, there is an urgent need to search for organic fertilizers that can enhance Chakhao rice growth.
In our study, plant growth promoting rhizobacteria (PGPR) were isolated from the rhizospheric soils of 6 Chakhao rice cultivars cultivated at different locations of Manipur, India. Even though there are many reports on the nutritional value of Chakhao rice and its importance to the people of Manipur, the application of PGPRs in Chakhao cultivation is still an underexplored area.
In the present study, 4 multi-trait PGP rhizospheric strains viz.CR12 (Bacillus subtilis strain CR12), CW11 (Bacillus paralicheniformis strain CW11), CA2 (Bacillus sp. CCMB1014 strain CA2) and CP2 (Bacillus licheniformis strain CP2) were chosen as potent biocontrol and biofertilizing agents as they can inhibit all the 5 fungal pathogens tested and were positive for all the PGP traits. The shortlisted potent strains exhibited antifungal activities against the 5 test fungal pathogens and showed significant mycelial growth inhibition against the fungal pathogens. Bacillus amyloquefaciens strain Bk7, a PGPR isolated from rice rhizosphere could inhibit the mycelial growth of Magnaporthe oryzae, Rhizoctonia solani, Botrytis cinerea and Fusarium graminearum36. Bacillus spp. strains B-1, B-2, B-3 and B-4 isolated from rice rhizospheric soils of Odisha, India showed strong antifungal activity (upto 90%) against the phytopathogens like Rhizoctonia solani, Sclerotium rolfsii and Sclerotium oryzae invitro37. A recent report showed a rice rhizospheric antifungal Bacillus sp. KM5 synthesized prominent antibiotics such as Bacillomycin F, Bacillomycin D, Bacillopeptin A, etc38. Significant inhibition of the five (5) fungal pathogens by the potent shortlisted strains can also be noted. Therefore they may hold potential to be developed as antifungal agents for rice cultivation.
Bacillus spp. strains K3 and N4 isolated from rice rhizospheric soil samples from different villages of Bardoli, Gujarat, India were reported to exhibit PGP activities like phosphate solubilization, IAA and siderophore production39. Phosphate solubilizing rhizobacteria (PSRB) such as Pseudomonas aeruginosa, Bacillus subtilis strain 1, Bacillus subtilis strain 2 and Bacillus subtilis strain 3, isolated from rice rhizosphere have been reported as promising agents for rice cultivation in soils deficient in phosphorus40. A rhizobacterial isolate UPMB19, belonging to the genera Lysinibacillus obtained from rice rhizosphere has been reported as a PGPR showing IAA and siderophore production and phosphate solubilization which can promote rice seed growth significantly41. ACC deaminase producing Bacillus sp. strain SB1-ACC3 isolated from the coastal rice field soils of West Bengal, India promoted rice seed germination42. Streptomyces corchorusii strain UCR3-16, obtained from rice rhizospheric soils showed antifungal activities, found positive for plant growth promoting traits such as IAA, ammonia, siderophore, ACC deaminase production and phosphate solubilization and the strain showed significant increase in growth and grain yield of rice plants27.The strains (CR12, CW11, CA2 and CP2) can produce IAA, siderophore, ammonia, ACC deaminase and solubilize phosphate. The shortlisted potent strains have the potential of plant growth promoting traits.
Strain CR12 exhibited production of phytohormone IAA in significant amount. It produced 73.83 µg/mL IAA on 10th day of incubation. A rhizospheric Bacillus sp. BPR7 isolated from common bean produced 17 µg/mL of IAA in 24 hrs incubation43. Goswami et al. (2014) has reported IAA production by PGPR from saline desert of Kutch in the range of 25 µg/mL of IAA or more after 72 hrs of incubation44. Strain CA2 could solubilize phosphate significantly (200 µg/mL) with a corresponding drop in pH (2.69) on the 3rd day of incubation. Bacillus sp. BPR7 isolated from common bean rhizosphere could solubilize upto 23 µg/mL of TCP (tricalcium phosphate) on the 7th day of incubation43. Yu et al. (2011) has reported several phosphate solubilizing bacterial (PSB) strains, from Walnut could solubilize phosphate in the range of 81.09 mg/L – 233.35 mg/L45. The effects of PSB on the growth of several crop plants have been reported by several workers who conducted their research under both greenhouse and limited field trial experiments46-47. Strain CA2 could produce siderophore in significant amount (74.37%) in SCNB. A common bean rhizospheric Bacillus sp. BPR7 could produce siderophore upto 38 µg/mL in 72 hrs43. The application of siderophore producing rhizobacteria as plant growth promoters have been documented by various researchers48-49.
Chakhao rice seedling germination by CR12, CW11, CA2 and CP2 were performed and exhibited higher vigor indices over the control. Vigor indices of CR12 (668.00), CW11 (478.91), CA2 (618.66) and CP2 (510.92) were found to be higher than vigor index of control (164.48). The PGPR strains UPMB10 (Bacillus sphaericus), Rhizobium strains (SB16, UPMR1006 and UPMR1102) reported by Mia et al. (2012) showed significant increase in seedling vigor and overall root growth of lowland rice variety MR21950. Bacillus subtilis strain MBI600, a PGPR obtained from Department of Entomology and Plant Pathology, Auburn University, AL, USA enhanced seedling vigor index of rice (13192) relative to the control (5028)51.
Conclusion
The best approach for sustainable Chakhao rice agriculture will be the use of microbes especially rhizospheric bacteria associated with the host plants. In the present study, 4 most promising rhizospheric bacteria from 6 different cultivars of Chakhao with PGP potential were found. All 4 shortlisted strains showed good inhibition against major rice fungal pathogens and so they can be used as promising antifungal agents. Moreover, these isolates also showed significant PGP activities. Then shortlisted strains showed notable higher seedling vigor indices over the control under in vitro conditions. Further studies are underway to investigate and confirm their growth promotion potency and biocontrol efficacy under pot trial and limited field conditions.
Acknowledgements
The authors gratefully acknowledge the Department of Biotechnology, GOI for the DBT-State Biotech Hub Grant (DBT-SBTHub) (BT/04/NE/2009), Department of Biochemistry, Manipur University, for extending infrastructural help that facilitated this research work, and also the University Grant Commission (UGC), Government of India for offering the UGC JRF/SRF fellowship to Sajida Sultana1.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
There is no funding sources
References
- Devi G.S., Shijagurumayum S., Singh C.B. Chakhao: Scented traditional rice of Manipur (India). Journal of Advanced Scientific Research. 2021;12(01):1-9.
- Wang Q., Han P., Zhang M., Xia M., Zhu H., Ma J., Hou M., Tang Z., Ling W. Supplementation of black rice pigment fraction improves antioxidant and anti-inflammatory status in patients with coronary heart disease. Asia Pac J Clin Nutr. 2007;16(1):295-301.
- Hui C., Bin Y., Xiaoping Y., Long Y., Chunye C., Mantian M., Wenhua L. Anticancer activities of an anthocyanin-rich extract from black rice against breast cancer cells in vitro and in vivo. Nutr Cancer. 2010;62(8):1128-1136. doi:10.1080/01635581.2010.494821
- Das K.R., Medhabati K., Nongalleima K., Devi H.S. The potential of dark purple scented rice-from staple food to nutraceutical. Curr World Environ. 2014;9(3):867-76.
- Sharma N., Singhvi R. Effects of chemical fertilizers and pesticides on human health and environment: a review. International journal of agriculture, environment and biotechnology. 2017;10(6):675-680.
- Hayat S., Faraz A., Faizan M. Root exudates: composition and impact on plant–microbe interaction. Biofilms in Plant and Soil Health. John Wiley & Sons Ltd. 2017;179-193. doi:10.1002/9781119246329
- Peng J., Ma J., Wei X., Zhang C., Jia N., Wang X., Wang E.T., Hu D., Wang Z. Accumulation of beneficial bacteria in the rhizosphere of maize (Zea mays L.) grown in a saline soil in responding to a consortium of plant growth promoting rhizobacteria. Annals of Microbiology. 2021;71(1):1-2
- Agrillo B., Mirino S., Tatè R., Gratino L., Gogliettino M., Cocca E., Tabli N., Nabti E., Palmieri G. An alternative biocontrol agent of soil-borne phytopathogens: A new antifungal compound produced Apr 1;221:60-9.by a plant growth promoting bacterium isolated from North Algeria. Microbiol Res. 2019
- Ramanathan A., Shanmugam V., Raguchander T., Samiyappan R. Induction of systemic resistance in Ragi against blast disease by Pseudomonas fluorescens. Ann PC Prot Soc. 2002;10(2):313-318.
- Ningthoujam D., Chanu S., Tamreihao K., Lynda R., Devi K., Jeeniita N. Plant growth promotion and biocontrol potential of a Streptomyces sp. Strain N3-3b isolated from the rhizosphere of chakhao, a black rice variety of manipur, India. Br Microbiol Res J. 2016;16(2):1-11. doi:10.9734/bmrj/2016/27422
- Singh P.P., Shin Y.C., Park C.S., Chung Y.R. Biological control of fusarium wilt of cucumber by chitinolytic bacteria. Phytopathology. 1999;89(1):92-99. doi:10.1094/PHYTO.1999.89.1.92
- Gopalakrishnan S., Pande S., Sharma M., Humayun P., Kiran B.K., Sandeep D., Vidya M.S., Deepthi K., Rupela O. Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Prot. 2011;30(8):1070-1078. doi:10.1016/j.cropro.2011.03.006
- Jog R., Nareshkumar G., Rajkumar S. Plant growth promoting potential and soil enzyme production of the most abundant Streptomyces spp. from wheat rhizosphere. J Appl Microbiol. 2012;113(5):1154-1164. doi:10.1111/j.1365-2672.2012.05417.
- Sadeghi A., Karimi E., Dahaji P.A., Javid M.G., Dalvand Y., Askari H. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J Microbiol Biotechnol. 2012;28(4):1503-1509. doi:10.1007/s11274-011-0952-7
- Passari A.K., Mishra V.K., Gupta V.K., Yadav M.K., Saikia R., Singh B.P. In vitro and in vivo plant growth promoting activities and DNA fingerprinting of antagonistic endophytic Actinomycetes associates with medicinal plants. PLoS One. 2015;10(9):e0139468. doi:10.1371/journal.pone.0139468
- Qin S., Miao Q., Feng W.W., Wang Y., Zhu X., Xing K., Jiang J.H. Biodiversity and plant growth promoting traits of culturable endophytic actinobacteria associated with Jatropha curcas L. growing in Panxi dry-hot valley soil. Appl Soil Ecol. 2015;93:47-55. doi:10.1016/j.apsoil.2015.04.004
- Asem I.D., Nongthombam A., Shaheen K., Heisnam N.D., Yurembam R., Asem R., Devi L.J., Khunjamayum R., Chanu T.P., Mukherjee S., Singh E.S.. Phenotypic characterization, genetic variability and correlation studies among ten Chakhao (scented) rice of Manipur. International Journal of Current Microbiology and Applied Sciences. 2019;8(2):612-8.
- Krieg N.R., Staley J.T., Brown D.R. Bergey’s Manual® of Systematic Bacteriology. Springer; 2010.
- Trivedi P., Pandey A., Palni L.M.S. In vitro evaluation of antagonistic properties of Pseudomonas corrugata. Microbiol Res. 2008;163(3):329-336. doi:10.1016/j.micres.2006.06.007
- Khamna S., Yokota A., Lumyong S. Actinomycetes isolated from medicinal plant rhizosphere soils: diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J Microbiol Biotechnol. 2009;25(4):649-655. doi:10.1007/s11274-008-9933-x
- Bano N., Musarrat J. Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Curr Microbiol. 2003;46(5):324-328. doi:10.1007/s00284-002-3857-8
- Mehta S., Nautiyal C.S. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr Microbiol. 2001;43(1):51-56. doi:10.1007/s002840010259
- Kapri A., Tewari L. Phosphate solubilization potential and phosphatase activity of rhizospheric Trichoderma spp.Brazilian Journal of Microbiology. 2010;41:787-795. doi:10.1590/S1517-83822010005000001
- Fiske C.H., Subbarow Y. The colorimetric determination of phosphorus. biol. Chem. 1925;66(2):375-400. doi:10.1016/S0021-9258(18)84756-1
- You J.L., Cao L.X., Liu G.F., Zhou S.N., Tan H.M., Lin Y.C. Isolation and characterization of actinomycetes antagonistic to pathogenic Vibrio spp. from nearshore marine sediments. World J Microbiol Biotechnol. 2005;21(5):679-682. doi:10.1007/s11274-004-3851-3
- Payne S.M. Detection, isolation, and characterization of siderophores. Methods in enzymology. 1994;235:329-344. org/10.1016/0076-6879(94)35151-1
- Sayyed R.Z., Badgujar M.D., Sonawane H.M., Mhaske M.M., Chincholkar S.B. Production of microbial iron chelators (siderophores) by fluorescent Pseudomonads. 2005. http://nopr.niscpr.res.in/handle/123456789/5776
- Cappucino J.C., Sherman N. Microbiology: A Laboratory Manual. Benjamin/Cumming Pub. Co; 1992.
- Dworkin M., Foster J.W. Experiments with some microorganisms which utilize ethane and hydrogen1. J Bacteriol. 1958;75(5):592-603. doi:10.1128/jb.75.5.592-603.1958
- El-Tarabily K.A. Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycete actinomycetes. Plant Soil. 2008;308(1-2):161-174. doi:10.1007/s11104-008-9616-2
- Tamreihao K., Ningthoujam D.S., Nimaichand S., Singh E.S., Reena P., Singh S.H., Nongthomba U. Biocontrol and plant growth promoting activities of a Streptomyces corchorusii strain UCR3-16 and preparation of powder formulation for application as biofertilizer agents for rice plant. Microbiol Res. 2016;192:260-270. doi:10.1016/j.micres.2016.08.005
- Abdul-Baki A.A., Anderson J.D. Vigor determination in soybean seed by multiple criteria 1. Crop Sci. 1973;13(6):630-633. doi:10.2135/cropsci1973.0011183x001300060013x
- Magray M.S.U.D., Kumar A., Rawat A.K., Srivastava S. Identification of Escherichia coli through analysis of 16S rRNA and 16S-23S rRNA internal transcribed spacer region sequences. Bioinformation. 2011;6(10):370-371. doi:10.6026/97320630006370
- Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I. GenBank. Nucleic Acids Res. 2020;48(D1):D84-D86. doi:10.1093/nar/gkz956
- Borah N., Athokpam F.D., Semwal R.L., Garkoti S.C. Chakhao (Black Rice; Oryza sativa L.): A culturally important and stress tolerant traditional rice variety of Manipur. Ind J Tradit Knowl. Published online 2018. Accessed February 27, 2023. http://nopr.niscpr.res.in/handle/123456789/45052
- Kakar K.U., Nawaz Z., Cui Z., Almoneafy A.A., Ullah R., Shu Q.Y. Rhizosphere-associated Alcaligenes and Bacillus strains that induce resistance against blast and sheath blight diseases, enhance plant growth and improve mineral content in rice. J Appl Microbiol. 2018;124(3):779-796. doi:10.1111/jam.13678
- Sethi S.K., Mukherjee A.K. Screening of biocontrol potential of indigenous Bacillus spp. isolated from rice rhizosphere against, solani, S. oryzae, S. rolfsii and response towards growth of rice. J Pure Appl Microbiol. 2018;12:41-53.
- Majumdar K., Chakraborty S. A new antifungal antibiotic from Bacillus sp. KM5 isolated from rice rhizospheric soil. Proc Natl Acad Sci India Sect B Biol Sci. 2019;89(1):333-344. doi:10.1007/s40011-017-0944-y
- Modi K., Patel P., Parmar K. Isolation, screening and characterization of PGPR from rhizosphere of rice. International Journal of Pure & Applied Biosciences. 2017;5(3):264-270.
- Gupta R., Kumari A., Sharma S., Alzahrani O.M., Noureldeen A., Darwish H. Identification, characterization and optimization of phosphate solubilizing rhizobacteria (PSRB) from rice rhizosphere. Saudi J Biol Sci. 2022;29(1):35-42. doi:10.1016/j.sjbs.2021.09.075
- Tan K.Z., Radziah O., Halimi M.S., Khairuddin A.R., Habib S.H., Shamsuddin Z.H. Rhizobacteria P. Isolation and characterization of rhizobia and plant growth-promoting rhizobacteria and their effects on growth of rice seedlings. American Journal of Agricultural and Biological Sciences. 2014;9(3):342-360.
- Bal H.B., Nayak L., Das S., Adhya T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil. 2013;366(1-2):93-105. doi:10.1007/s11104-012-1402-5
- Kumar P., Dubey R.C., Maheshwari D.K. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res. 2012;167:493-499. org/10.1016/j.micres.2012.05.002
- Goswami D., Dhandhukia P., Patel P., Thakker J.N. Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis Microbiol Res. 2014;169(1):66-75. doi.org/10.1016/j.micres.2013.07.004
- Yu X., Liu X., Zhu T.H., Liu G.H., Mao C. Isolation and characterization of phosphate-solubilizing bacteria from walnut and their effect on growth and phosphorus mobilization. Biology and Fertility of Soils, 2011;47(4):437-446.
- Hariprasad P., Niranjana S.R. Isolation and characterization of phosphate solubilizing rhizobacteria to improve plant health of tomato. Plant and soil. 2009;316(1):13-24.
- Pande A., Pandey P., Mehra S., Singh M., Kaushik S. Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. Journal of Genetic Engineering and Biotechnology.2017;15(2):379-391. org/10.1016/j.jgeb.2017.06.005
- Ghavami N., Alikhani H.A., Pourbabaei A.A., Besharati H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. Journal of Plant Nutrition.2017;40(5):736-746.
- Kumar P., Thakur S., Dhingra G.K., Singh A., Pal M.K., Harshvardhan K., Dubey R.C., Maheshwari D.K. Inoculation of siderophore producing rhizobacteria and their consortium for growth enhancement of wheat plant. Biocatalysis and agricultural biotechnology.2018;15:264-269. doi:10.1016/j.bcab.2018.06.019
- Mia M.B., Shamsuddin Z.H., Mahmood M. Effects of rhizobia and plant growth promoting bacteria inoculation on germination and seedling vigor of lowland rice. African Journal of Biotechnology. 2012;11(16):3758-3765.
- Kumar K.V.K., Reddy M.S., Kloepper J.W., Lawrence K.S., Yellareddygari S.KR., Zhou X.G., Sudini H., Reddy E.C.S., Groth D.E., Miller M.E. Screening and selection of elite plant growth promoting rhizobacteria (PGPR) for suppression of Rhizoctonia solani and enhancement of rice seedling vigor. J Pure Appl Microbiol. 2011;5(2):1-11.

