Introduction
Today’s global scenario presents many constraints to agricultural production, leading to a decline in soil and soil fertility as well as a decline in agricultural output in general. Things are even worse for resource-poor farmers, who cannot afford to use high doses of organic fertilizers for crop production 1. By 2020, 28.8 million tons of nutrients are needed to produce million tons (approximately 321 million) of grain. However, available nutrients were 21.6 million tons – a wide gap of 7.2 million tons between nutrient supply and nutrient removal. 2 In addition to reducing urea-N and preventing soil depletion, biological nitrogen fixation (BNF) technology also considerably lowers environmental pollution.3 Since they are beneficial to the environment and economical for farmers, organic fertilisers are seen to be a key component in productivity, soil sustainability, and environmental protection 4. Farmers may gain financially from using this organic fertiliser, which also assures continued food production. The worldwide green revolution will begin with the application of chemical-free fertilisers or microbial inoculation.5
Bio-fertilizers are mainly live microbial products including bacteria, algae and fungi, singly or in combination with the effect of improving nutrients available in the soil for plants. Essential nutrients change from unusable to usable form with the assistant of microbial activities; activities also include phosphate solubilization, nitrogen fixation, soil biodegradation and plant growth hormone secretion. Bio-fertilizers are ecological, high-yielding, easier for farmers to use and more efficient.2
Azospirillium species
The genera Azotobacter and Azospirillium increase the yield of cereals and legumes under field conditions. 4 Azospirillium is solitary of the earliest identified/discovered and best-characterized plants that promote rhizobacteria covering tropical and temperate regions commonly applied to grass, rice, wheat, sugarcane and many other crops. 6. It grows profusely at pH-7 and can form spores in acidic soils at pH 4.8. Azosprilllium is a motile, non-fermenting, vibrioid, gram-negative nitrogen-fixing rhizome. It is an encapsulated microcapsule; Sheath are composed of a network of inverse polysaccharides ensuring stress tolerance and increased shelf life. Azospirillium is an α subgenus of proteobacteria with the original name Spirillum lipoferumisolated by Beijerinck from the Netherlands Laterly, by Schroder in 1932 from the soil of Germany and Austria7. The organism showed increased nitrogen content in the N2-free medium. Azospirillium can grow aerobically and anaerobically in different geographical climates. It uses organic acids on sugars as carbon sources, ammonium or nitrates as nitrogen sources, and it can fix nitrogen under microscopic and free living conditions8. A number of processes, including as the production and release of amino acids, IAA (indole acetic acid), Cytokines, Gibberellins, and other polyamines, as well as root development and the subsequent increase in water and nutrient intake, all work to promote Azospirillium. A huge genus, Azospirillium, has around 113 plant species from 35 different plant families. 9-10 Analyzed the results of multiple field trials with different non-legume crops, representing more than 20 years worldwide in a variety of weather and soil conditions, examining that 30% yield increase can reach 70% response time with Azospirillum inoculation. 11 reported that Azospirillum sp. inoculated on upland crops showed yield responses in winter cereals (14.0%), summer cereals (9.5%), and also in legumes (6.6%). 12 reported Azospirillum spp. isolated from Himalaya valley and other places from Lake Baiyang. The species isolated in 2019 were – A.palustre 13, A. griseum 14, A. Ramasamy 15, A.Agricola 16, A.soli 17, Nitrospirillum & Niveispirillum 18, A.Himalayense 19, A.fermentation 20, A.Humicireducens 21, A.formosense 22, A.thiophilum 23, A.palatum 24, A.picis 25, A .rugosum 24, A.canadense & A.zeae 26, A.melinis 27, A.oryzae 28, A.doeberinerae 29, A.largimobile 30, A.irakense 31, A. .halopraeferens 32, A.amazonense 33, A.lipoferum & A.brasilense 34.
Root attachment to surface and site of root colonization
Sites suitable for colony colonisation have been shown to be where Azospirillum colonisation occurs on the surface of roots. In the most studied plant species, the bacteria form aggregation colonies that are supported by enormous fibrous material around the tips of elongated hairs and roots. Two distinct phases of Azospirillum’s attachment to wheat have been examined by 35. Bacterial proteins rule the quick and weak adsorption phase. The second phase is more prolonged, active, and irreversible; it could be based on the extracellular surface polysaccharides of bacteria.
The country becomes more self-sufficient in food production thanks to the use of chemical fertilisers, but this practise damages the environment, has negative effects on human health, contaminates groundwater, and promotes soil acidification owing to a decline in organic matter. Owing to inadequate plant absorption of these chemical fertilisers, they enter water bodies through rains, create eutrophication there, and have an impact on aquatic life. Finding management practises that can impact plant development, a general improvement in root growth, significant bio-control activities, and numerous mechanisms of the plant-microorganism interplay will be a future issue.
Materials and Methods
In the present investigation, the Azospirillium isolates mentioned in Table 1 were derived from the rhizomes of different non-legumes collected from different districts of Saurashtra region.
Sample collection
The Isolates were collected at a depth of 5-6 cm from the Rhizome Zone of the Plants (Maize, Cotton, Jowar, Sugarcane, Bajra, Wheat, Groundnut etc.) stored at 4 ℃. Species were isolated from roots and soil by serial dilution.36
Root isolation
Roots, washed thoroughly with water, cut into small pieces and shaken in a shaker for 30 min in sterile distilled water; 0.1 ml of water was inoculated into NFB semi-solid medium and incubated for 5-7 days at 30-35 °C to form a white film 37,38.
Soil Isolation
1 g soil was diluted in order to 10-6 with sterile distilled water. 1 ml of 10-4 to 10-6 diluent was incubated in tubes containing semi-solid NFB medium (Nitrogen-Free Bromothymol) and incubated for 5-7 days at 30-35°C to form films. These films have demonstrated positive results for the development of Azospirillium. Colonies were spread on Petri dishes containing solid NFB medium (Bromothymol without Nitrogen) and incubated at 30-35 °C for 72 h to isolate single colonies39.
Media Formulation
NFB medium contains 5 g malic acid, 4.0 g sodium chloride, 0.5 g potassium phosphate, 0.05 g ferric sulfate, 0.01 g manganese sulfate, 0.10 g magnesium sulfate, 0.02 g sodium chloride, 0.01 g calcium chloride, 0.002 g sodium molybdate dihydrate, 1000 ml water distillation, 2.0 ml bromothymol blue (BTB) – (0.5% alcohol solution) Final pH 6.6 to 6.8, 1.75 g of semi-solid medium and 15 g of solid medium 40,41.
Morphology and Classification
The juvenile inoculum from the semi-solid medium was applied on a clean slide, thermostated, then flooded with iodine for 1 minute, decolored with ethanol 95% in a matter of seconds, gently rinsed with tap water, and cleaned with Safranin stains. For viewing with an oil immersion lens, slides were air-dried 42. Gram-negative, rod-, spiral-, spiral-, and filamentous bacteria were seen under a microscope. The physical characteristics of the Azospirillium isolates are listed in Table 2.
Biochemical Characteristics
According to Bergey’s Guide to Determining Bacteria, 9th Edition, isolates were characterized for different biochemical assays. The tests were carbohydrate fermentation, catalase test, mobility test, citrate utilization, triple sugar iron, indole production, MR-VP test, peptone nitrate reduction, ammonia production, urea hydrolysis, gelatin hydrolysis and lipid hydrolysis 3,43.
Production of Phytohormones
24 hour old explants were allowed to develop in tryptophan culture at 30 °C for 24 hours in order to measure the quantity of Indole Acetic Acid (IAA) produced in each isolate using Salkowski’s reagent. After incubation, the broth was centrifuged, and Salkowski reagent 2 ml were combined with 1 ml of the supernatant. IAA generation is shown by the colour shift and absorption at 540 nanometers. Before to using the set in potted culture studies, each isolate was maintained in three sets 44.
Results and Discussion
In this present study, 50 isolates were isolated from different Rhizome Regions of the roots and soil samples. Samples were taken from 10 districts of Saurashtra region – Rajkot 19, Junagadh 9, Amerali 5, Botad 5, Morbi 2, Dwarka 2, Kutch 3, After Ahemdabad 2, Surat 2 and Vadodara 1 are presented in Table 1
Characteristics of the Colony
All 50 isolates were morphologically classified by Gram staining based on their characteristics such as shape, size, margin texture, elevation, and opacity. Of these isolates, 44 strains of bacteria were gram-negative, all of which were circular in shape with an entire margin and smooth texture, followed by a predominantly small, but some are medium sized and slightly raised or flat elevation. 14 colonies were opaque white, 32 colonies were transparent and 4 colonies were translucent. 6 Strain of bacteria were Gram Positive.
Biochemical characteristics
Models: – S-2, S-3, S-7, S-11, S-12, S-13, S-15, S-16, S-17, S-21, S-27, S- 29, S-30, S-41, S-46 are portable Fermenting sugars such as glucose, fructose, lactose, mannitol, it hydrolyzes gelatin, and urea, they can also produce H2S, indol and ammonia. The results are shown in Table 3. These isolates were then screened for potted culture experiments.
Production of Phytohormones
Salkowski reagent was used to assess the IAA generation of 50 isolates at 540 nm. Figure 4 displays the three isolates that produced the greatest IAA, S-27 190.19 g/ml, S-46 195.20 g/ml, and S-21 90.04 g/ml. The histogram’s X-axis and l-Y axis both display the number of isolates, shows the amount of IAA created by Isolate.
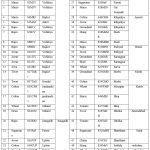 |
Table 1: shows isolates from Saurashtra region. |
*Sample– Plant sample, Sample Code S- Sample, No.- Sample number, Plant Initials, Plant Numbers and City/Village Initials, City/Village-Sample collected, District
Table 2: Colony Characteristic of different isolates.
| Sample | +/- | Shape | Size | Margin | Texture | Elevation | Opacity |
| S1 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S2 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S3 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S5 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S6 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S7 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S9 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S10 | -ve | Round | Medium | Entire | Smooth | Raised | Transperant |
| S11 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S12 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S13 | -ve | Round | Small | Entire | Smooth | Flat | Opaque |
| S14 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S15 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S16 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S18 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S19 | -ve | Round | Small | Entire | Smooth | Raised | Opaque |
| S20 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S21 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S22 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S23 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S24 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S26 | -ve | Round | Medium | Entire | Smooth | Raised | Transperant |
| S27 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S28 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S29 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S30 | -ve | Round | Small | Entire | Smooth | Raised | Opaque |
| S31 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S32 | -ve | Round | Small | Entire | Smooth | Raised | Transperant |
| S33 | -ve | Round | Small | Entire | Smooth | Raised | Transperant |
| S34 | -ve | Round | Small | Entire | Smooth | Flat | Opaque |
| S35 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S36 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S37 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S39 | -ve | Round | Small | Entire | Smooth | Raised | Transperant |
| S40 | -ve | Round | Medium | Entire | Smooth | Raised | Translucent |
| S41 | -ve | Round | Medium | Entire | Smooth | Raised | Transperant |
| S42 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S43 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S44 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S46 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S47 | -ve | Round | Small | Entire | Smooth | Slight Raised | Transperant |
| S48 | -ve | Round | Small | Entire | Smooth | Slight Raised | Opaque |
| S49 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
| S50 | -ve | Round | Small | Entire | Smooth | Flat | Transperant |
* S-Sample number, +ve – Gram Positive Bacteria and –ve – Gram Negative Bacteria
Table 3: Shows Biochemical Characterization of different Isolates
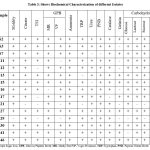 |
Table 3: Shows Biochemical Characterization of different Isolates |
*TSI- Triple Sugar Iron, GPB- Glucose Phosphate Broth, MR- Methyl Red VP- Voges Proskauer, TRP-Tryptophan, PNB- Peptone Nitrate Broth
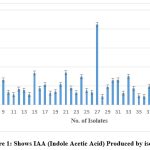 |
Figure 1: Shows IAA (Indole Acetic Acid) Produced by isolates |
Discussion
One of the most well researched plant growth-promoting bacteria, Azospirillum, is found specifically in the rhizomes and intercellular regions of cereal roots and other plant roots.43 The favourable benefits of this bacterium on plant development have been demonstrated through a number of pathways indicated by plant-Azospirillum interactions. The concept of the multiple mechanism hypothesis was developed through the study of numerous phytohormones, plant regulators, nitrogen fixation, phosphate solubilizers, a wide range of molecules and enzymes, enhanced membrane activity, root system proliferation, increased water and mineral absorption, reduced pathogens, environmental stressors, and competition against pathogens. 12 The most significant agricultural crop in the world is wheat, which is farmed in a variety of habitats, from marshlands to desert areas. Since the 1970s, scientists have been examining the results of Azospirillum inoculating grass. 45 Azospirillum, one of 113 species in 35 plant families, is a soil bacterium that encourages plant development. 8 The number of primary root nodules dramatically increased when A. brasilense, an efficient generator of IAA, was applied to alfalfa seeds; this increase was associated with inoculum size. 11 One isolation (Sp1) and the second isolate (Sp2) revealed 99% similarity to Azospirillum brasilense and Azospirillum zeae, respectively, based on 16S rRNA sequence analysis. In a pot experiment, the impact of these two isolates on bread wheat was investigated. 46 The greatest obstacle to effective inoculation is the toxicity of pesticides employed in seed treatments. In order to avoid direct bacterial exposure to pesticides, we investigated different approaches to inoculating maize and wheat seeds in this work.47
Conclusion
The current study includes 50 isolates from the Saurashtra Area, of which 2 isolates, S-27 and S-46, were shown to produce the greatest IAA (Indole Acetic Acid) production when tested against Salkowski’s reagent. These 2 isolates will be examined for 16s rRNA sequencing as well as evaluated against pot cultures. In order to create bio-fertilizers and field characteristics, it can be employed. The Saurahtra area would be a better place to isolate species of Azospirillium sp., which might help improve numerous growth indices, mineral uptake, and water absorption.
Acknowledgment
The Authors are thankful to the Head of the Department of Microbiology and Biotechnology, Atmiya University, Rajkot for providing suggestion, infrastructure and facilities
Funding Sources
Sarvangi Rabara is thankful to Government of Gujarat for providing financial assistance under its ‘SHODH SCHEME’.
References
- Khanna, R., Pawar, J., Gupta, S., Verma, H., Trivedi, H., Kumar, P., & Kumar, R. (2019). Efficiency of biofertilizers in increasing the production potential of cereals and pulses: A. Journal of Pharmacognosy and Phytochemistry, 8(2), 183-188.
- Kalsoom, M., Rehman, F. U., Shafique, T. A. L. H. A., Junaid, S. A. N. W. A. L., Khalid, N., Adnan, M., … & Ali, H. (2020). Biological importance of microbes in agriculture, food and pharmaceutical industry: A review. Innovare Journal Life Sciences, 8(6), 1-4.
CrossRef - Shamim, M. I. A., DIJKSTRA, F. A., ABUYUSUF, M., & HOSSAIN, A. I. (2015). Synergistic effects of biochar and NPK fertilizer on soybean yield in an alkaline soil. Pedosphere, 25(5), 713-719.
CrossRef - Mohammadi, K., & Sohrabi, Y. (2012). Bacterial biofertilizers for sustainable crop production: a review. ARPN J Agric Biol Sci, 7(5), 307-316.
- Fukami, J., Cerezini, P., & Hungria, M. (2018). Azospirillum: benefits that go far beyond biological nitrogen fixation. Amb Express, 8(1), 1-12.
CrossRef - Arulselvi, T., Kanimozhi, G., & Panneerselvam, A. (2018). Isolation and Characterization of Phosphate Solubilizing Fungi from the Soil Sample of MUTHUPET Mangroves. International Journal of Scientific Research in Science and Technology, 4(9), 30-37.
- Narayan, R., & Gupta, N. C. (2018). Isolation, cultural and physiological characterisation of Azospirillum from acidic soils of Ranchi. J Pharmacognosy and Phytochem, 7(4), 2611-2617.
CrossRef - O’Neal, L., Ryu, M. H., Gomelsky, M., & Alexandre, G. (2017). Optogenetic manipulation of cyclic di-GMP (c-di-GMP) levels reveals the role of c-di-GMP in regulating aerotaxis receptor activity in Azospirillum brasilense. Journal of bacteriology, 199(18), e00020-17.
CrossRef - Zeffa, D. M., Perini, L. J., Silva, M. B., de Sousa, N. V., Scapim, C. A., Oliveira, A. L. M. D., … & Azeredo Goncalves, L. S. (2019). Azospirillum brasilense promotes increases in growth and nitrogen use efficiency of maize genotypes. Plos one, 14(4), e0215332.
CrossRef - Galindo, F. S., Rodrigues, W. L., Biagini, A. L. C., Fernandes, G. C., Baratella, E. B., da Silva Junior, C. A., … & Teixeira Filho, M. C. M. (2019). Assessing forms of application of Azospirillum brasilense associated with silicon use on wheat. Agronomy, 9(11), 678.
CrossRef - Cassán, F., & Diaz-Zorita, M. (2016). Azospirillum sp. in current agriculture: From the laboratory to the field. Soil Biology and Biochemistry, 103, 117-130.
CrossRef - Cassán, F., Coniglio, A., López, G., Molina, R., Nievas, S., de Carlan, C. L. N., … & Mora, V. (2020). Everything you must know about Azospirillum and its impact on agriculture and beyond. Biology and Fertility of Soils, 56(4), 461-479.
CrossRef - Tikhonova, E. N., Grouzdev, D. S., & Kravchenko, I. K. (2019). Azospirillum palustre sp. nov., a methylotrophic nitrogen-fixing species isolated from raised bog. International Journal of Systematic and Evolutionary Microbiology, 69(9), 2787-2793.
CrossRef - Yang, Y., Zhang, R., Feng, J., Wang, C., & Chen, J. (2019). Azospirillum griseum sp. nov., isolated from lakewater. International Journal of Systematic and Evolutionary Microbiology, 69(12), 3676-3681.
CrossRef - Anandham, R., Heo, J., Krishnamoorthy, R., Senthil Kumar, M., Gopal, N. O., Kim, S. J., & Kwon, S. W. (2019). Azospirillum ramasamyi sp. nov., a novel diazotrophic bacterium isolated from fermented bovine products. International journal of systematic and evolutionary microbiology, 69(5), 1369- 1375.
CrossRef - Lin, S. Y., Liu, Y. C., Hameed, A., Hsu, Y. H., Huang, H. I., Lai, W. A., & Young, C. C. (2016). Azospirillum agricola sp. nov., a nitrogen-fixing species isolated from cultivated soil. International journal of systematic and evolutionary microbiology, 66(3), 1453-1458.
CrossRef - Lin, S. Y., Hameed, A., Liu, Y. C., Hsu, Y. H., Lai, W. A., Shen, F. T., & Young, C. C. (2015). Azospirillum soli sp. nov., a nitrogen-fixing species isolated from agricultural soil. International journal of systematic and evolutionary microbiology, 65(Pt_12), 4601-4607.
CrossRef - Lin, S. Y., Hameed, A., Shen, F. T., Liu, Y. C., Hsu, Y. H., Shahina, M., … & Young, C. C. (2014). Description of Niveispirillum fermenti gen. nov., sp. nov., isolated from a fermentor in Taiwan, transfer of Azospirillum irakense (1989) as Niveispirillum irakense comb. nov., and reclassification of Azospirillum amazonense (1983) as Nitrospirillum amazonense gen. nov. Antonie van Leeuwenhoek, 105(6), 1149-1162.
CrossRef - Tyagi, S., & Singh, D. K. (2014). Azospirillum himalayense sp. nov., a nifH bacterium isolated from Himalayan valley soil, India. Annals of microbiology, 64(1), 259-266.
CrossRef - Lin, S. Y., Liu, Y. C., Hameed, A., Hsu, Y. H., Lai, W. A., Shen, F. T., & Young, C. C. (2013). Azospirillum fermentarium sp. nov., a nitrogen-fixing species isolated from a fermenter. International journal of systematic and evolutionary microbiology, 63(Pt_10), 3762-3768.
CrossRef - Zhou, S., Han, L., Wang, Y., Yang, G., Zhuang, L., & Hu, P. (2013). Azospirillum humicireducens sp. nov., a nitrogen-fixing bacterium isolated from a microbial fuel cell. International journal of systematic and evolutionary microbiology, 63(Pt_7), 2618- 2624.
CrossRef - Lin, S. Y., Shen, F. T., Young, L. S., Zhu, Z. L., Chen, W. M., & Young, C. C. (2012). Azospirillum formosense sp. nov., a diazotroph from agricultural soil. International journal of systematic and evolutionary microbiology, 62(Pt_5), 1185- 1190.
CrossRef - Lavrinenko, K., Chernousova, E., Gridneva, E., Dubinina, G., Akimov, V., Kuever, J., … & Grabovich, M. (2010). Azospirillum thiophilum sp. nov., a diazotrophic bacterium isolated from a sulfide spring. International journal of systematic and evolutionary microbiology, 60(12), 2832-2837.
CrossRef - Zhou, Y., Wei, W., Wang, X., Xu, L., & Lai, R. (2009). Azospirillum palatum sp. nov., isolated from forest soil in Zhejiang province, China. The Journal of general and applied microbiology, 55(1), 1-7.
CrossRef - Lin, S. Y., Young, C. C., Hupfer, H., Siering, C., Arun, A. B., Chen, W. M., … & Yassin, A. F. (2009). Azospirillum picis sp. nov., isolated from discarded tar. International journal of systematic and evolutionary microbiology, 59(4), 761-765.
CrossRef - Mehnaz, S., Weselowski, B., & Lazarovits, G. (2007). Azospirillum zeae sp. nov., a diazotrophic acterium isolated from rhizosphere soil of Zea mays. International journal of systematic and evolutionary microbiology, 57(12), 2805-2809.
- Peng, G., Wang, H., Zhang, G., Hou, W., Liu, Y., Wang, E. T., & Tan, Z. (2006). Azospirillum melinis sp. nov., a group of diazotrophs isolated from tropical molasses grass. International Journal of Systematic and Evolutionary Microbiology, 56(6), 1263-1271.
CrossRef - Xie, C. H., & Yokota, A. (2005). Azospirillum oryzae sp. nov., a nitrogen fixing bacterium isolated from the roots of the rice plant Oryza sativa. International journal of systematic and evolutionary microbiology, 55(4), 1435-1438.
CrossRef - Eckert, B., Weber, O. B., Kirchhof, G., Halbritter, A., Stoffels, M., & Hartmann, A. (2001). Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. International Journal of Systematic and Evolutionary Microbiology, 51(1), 17-26.
CrossRef - Dekhil, S. B., Cahill, M., Stackebrandt, E., & Sly, L. I. (1997). Transfer of Conglomeromonas largomobilis subsp. largomobilis to the genus Azospirillum as Azospirillum largomobile comb. nov., and elevation of Conglomeromonas largomobilis subsp. parooensis to the new type species of Conglomeromonas, Conglomeromonas Azospirillum (pp. 3-26). Springer, Cham.
- Khammas, K. M., Ageron, E., Grimont, P. A. D., & Kaiser, P. (1989). Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Research in microbiology, 140(9), 679-693.
CrossRef - Reinhold, B., Hurek, T., Fendrik, I., Pot, B., Gillis, M., Kersters, K., … & De Ley, J. (1987). Azospirillum halopraeferens sp. nov., a nitrogen- fixing organism associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth). International Journal of Systematic and Evolutionary Microbiology, 37(1), 43-51.
CrossRef - Falk, E. C., Döbereiner, J., Johnson, J. L., & Krieg, N. R. (1985). Deoxyribonucleic acid homology of Azospirillum amazonense Magalhães et al. 1984 and emendation of the description of the genus Azospirillum. International Journal of Systematic and Evolutionary Microbiology, 35(1), 117-118.
CrossRef - Tarrand, J. J., Krieg, N. R., & Döbereiner, J. (1978). A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Canadian journal of microbiology, 24(8), 967-980.
CrossRef - Holguin, G., & Bashan, Y. (1996). Nitrogen fixation by Azospirillum brasilense Cd is promoted when co-cultured with a mangrove rhizosphere bacterium (Staphylococcus sp.). Soil Biology and Biochemistry, 28(12), 1651-1660.
CrossRef - Sulaiman, K. H., Al-Barakah, F. N., Assafed, A. M., & Dar, B. A. M. (2019). Isolation and identification of Azospirillum and Azotobacter species from Acacia spp. at Riyadh, Saudi arabia. Bangladesh Journal of Botany, 48(2), 239-251.
CrossRef - Akbari, G. A., Arab, S. M., Alikhani, H. A., Allakdadi, I., & Arzanesh, M. H. (2007). Isolation and selection of indigenous Azospirillum spp. And the IAA of superior strains effects on wheat roots. World Journal of Agricultural Sciences, 3(4), 523-529.
- Topre, S. D., Panikar, S. S., Mahajani, S. U., & Patil, S. B. (2011). Biofertilizer: A novel approach for agriculture. Journal of Agricultural Biotechnology and Sustainable Development, 3(10), 205-208.
- Day, J. M., & Döbereiner, J. (1976). Physiological aspects of N2-fixation by a Spirillum from Digitaria roots. Soil Biology and Biochemistry, 8(1), 45-50. 39.
CrossRef - Reis, V. M., Baldani, V. L. D., & Baldani, J. I. (2015). Isolation, identification and biochemical characterization of Azospirillum spp. and other nitrogen-fixing bacteria. In Handbook for Azospirillum (pp. 3-26). Springer, Cham.
- Gandhimaniyan, K., Balamurugan, V., Ambedkar, G., Sabaridasan, A., Subramanian, M., & Anandababu, S. STUDIES ON THE ISOLATION AND CHARACTERIZATION OF Azospirillum SP. IN RHIZOSPHERE SOIL OF MAIZE.
- Mohamed, M. G., Mohamed, H. M., Gameh, M. A., & El-Rewainy, H. M. (2018). Isolation and Characterization of Azospirillum Isolates from Soil and Their Effect on Growth and Yield of Wheat (Triticum aestivum L.) under Different Levels of Nitrogen Fertilizer. Assiut Journal of Agricultural Sciences, 49(3), 107-116.
CrossRef - Hingole, S. S., & Pathak, A. P. (2013). Report on efficient salt stable Azospirillum a Lonar Soda Lake isolate. Science Research Reporter, 3(2), 200-203.
- Zeffa, D. M., Perini, L. J., Silva, M. B., de Sousa, N. V., Scapim, C. A., Oliveira, A. L. M. D., … & Azeredo Goncalves, L. S. (2019). Azospirillum brasilense promotes increases in growth and nitrogen use efficiency of maize genotypes. Plos one, 14(4), e0215332.
CrossRef - Alexandre, G. (2017). Azospirillum brasilense, a beneficial soil bacterium: isolation and cultivation. Current protocols in microbiology, 47(1), 3E-1.
CrossRef - Baldani, V. L. D., & Döbereiner, J. (1980). Host-plant specificity in the infection of cereals with Azospirillum spp.Soil biology and biochemistry, 12(4), 433-439.
CrossRef - Fukami, J., Nogueira, M. A., Araujo, R. S., & Hungria, M. (2016). Accessing inoculation methods of maize and wheat with Azospirillum brasilense. Amb Express, 6(1), 1-1
CrossRef

