Introduction
Over the past decades, heavy metals from agricultural, mining, sewage and other industrial practices are flooding the earth surface. This raises great concern as these metals such as cadmium, lead, mercury, zinc, copper, nickel, chromium and arsenic etc. are non-degradable by any physical or biological process resulting in long term threat to the environment. These metals are persistent in the soil for a long period and exert toxicity to the living entities interacting thereon. Remediation strategies based on physical and mechanical techniques include soil incineration, excavation and landfill, soil washing, solidification and electric field application1. These processes are disadvantageous in certain respect like their high cost, lesser efficiency in low concentration of heavy metals, creating irreversible changes to the properties of soil etc.
In this regard, bioremediation strategies are more environment-friendly, efficient and cost effective in mitigating heavy metal load in metal contaminated soil. Plant species offer excellent remediation effect as they populate naturally in soil ecosystem over a large area with minimum installation and maintenance cost. Moreover, plants are natural catalysts for building soil fertility, preventing soil-erosion and leaching of metal contaminants by stabilizing them. Metal accumulation, tolerance and bioavailability by plant species, on the other hand, are heavily dependent on the nature and intensity of rhizospheric, arbuscular and endophytic microbial population 2,3. Especially plant growth promoting rhizobateria (PGPR) are well known for promoting plant growth, metal tolerance and overall fitness of the plant. These bacteria are capable of synthesizing organic acids, antibiotics, enzymes, siderophores and various phytohormones which encourage plant root, shoot development, nutrient uptake and bioavailability of contaminating heavy metals 1,4.
Bacillus subtilis is a common PGPR which strengthen plant-microbe interaction by biofilm formation, efficiently reduce the release of nitrogen and ammonia gas, elicit induced systemic resistance, ameliorate abiotic stress by enzymatic and hormonal regulation and inhibit spreading of antibiotic resistance gene and transposons 5. These bacteria are potent biocontrol agents due to their lipopeptides. They also play prominent role in plant growth and soil improvement. In the present study, we report a Bacillus subtilis species which was isolated from heavy metal laden waste water collected from various points of the Green Belt Canal carrying waste materials released from Haldia Industrial Belt, West Bengal. This Industrial Belt is a hub of several chemical and petrochemical based industrial units like detergent manufacturing unit, pesticide manufacturing unit, lead-acid battery manufacturing unit, petroleum refinery unit, edible oil manufacturing unit and many others.
In order to characterize and determine the metal absorbing/stabilizing property of this microorganism, the present study was carried out using Indian mustard and tomato in a pot assay.
Materials and Methods
Collection of water samples and determination of pH
Water samples were collected from the Haldia Industrial Park (22° 4′ 0.228″ N and 88° 4′ 11.3124″ E) at eight points such as: the effluent release point near Indian Oil Corporation Ltd.(IOC) second Gate (sampling point 1), Indian Oil Corporation Ltd.(IOC) Main Gate (sampling point 2), Hooghly Met Coke Gate (sampling point 3), Patikhali Gate (sampling point 4), United Phosphorus Ltd. (UPL)Gate (sampling point 5), Indorama Plant(sampling point 6), Haldia Petrochemicals Ltd. (HPL) Gate (sampling point 7), and Naphtha Plant (sampling point 8) and transported to the authors’ laboratory for examination in sterile amber-coloured bottles. A portable digital pH meter (sensitivity 0.01) was used to determine the pH of the collected water samples.
Enumeration of heavy metal resistant bacteria from collected waste-laden water
Heavy metal resistant bacteria were isolated from collected water samples by spread plate technique using nutrient agar (containing 5g peptone, 3g beef extract, 15g agar and 5 g sodium chloride in 1000ml distilled water at pH 7.0) plates supplemented with 1000 ppm Pb(NO3)2. Water sample (0.1 ml) was spread on metal amended nutrient agar plates using a sterile spreader. The plates were incubated at 37°C in the incubator for 48 hours for bacterial growth to appear and then colonies were counted.
Selection of lead resistant bacteria from the resistant strains and determination of MIC
From the pool of resistant bacteria, lead resistant strains were opted as they were more populated. MIC of the resistant bacteria was determined using different concentration of lead in the metal augmented media like 1000 ppm, 1200 ppm, 1500 ppm and 2000ppm. At 37°C, the plates were incubated and the colonies were counted 6.
Isolation of pure culture of the lead resistant bacteria and its characterization
Out of eight waste water samples, five water samples were rich in lead resistant bacteria. Amongst them, most resistant strain was obtained from sampling point 7, which was isolated as single colony via quadrant streaking method using lead amended nutrient agar plate. Pure culture was stored in NA medium slants in vials under refrigerated conditions (4°C) before being tested further. This strain was further explored for it’s biological properties.
Determination of optimum pH and temperature for the resistant bacteria
Three 50 ml nutrient broths [Composition: 5 g peptone, 3 g beef or yeast extract, 5 g sodium chloride (NaCl), 1000 ml distilled water at pH 7.0] of different pH (4.0, 7.0, and 9.0) were prepared in a nephalometric flask by drop wise addition of HCl (to achieve pH-4.0) and NaOH (to achieve pH-7.0) to determine the optimum pH of the selected strain of lead resistant bacteria. Under aseptic conditions, the chosen strain was inoculated into them. These broths were shaken before incubation for 24 hours at 37°C. Turbidity was observed after 24 hours and measured in a colorimeter at 600nm, with the results being reported 7. For determination of optimum temperature, three 50 ml nutrient broths were prepared and the selected strain was inoculated into them under aseptic condition. These broths were incubated at different temperatures (25°C, 37°C, and 50°C) for 24 hours. Turbidity was observed after 24 hours and measured in a colorimeter at 600nm, with the results being reported 7.
Antibiotic susceptibility test
The resistant bacteria were tested for antibiotic susceptibility using the standard cup plate diffusion technique in Mueller Hinton agar (HiMedia, India). Penicillin, Ofloxacin, and Amoxyclav were among the antibiotics used. Pure culture was used as inoculum. The wells were prepared using a sterile borer and 0.1 ml culture was spread on Mueller Hinton agar (HiMedia, India) using a sterile spreader. Different antibiotic solutions of different concentrations (0.01mg/ml and 0.005 mg/ml) were poured into different wells using sterile micropipette. One plate was done as control. The zone of inhibition was measured after 24 hours and recorded in millimetres. According to the dose of antibiotics used and the zone of inhibition, the isolates were graded as highly resistant, moderately resistant, or susceptible 7.
Gram Staining of the pure culture
Gram nature of the resistant pure culture was determined by Gram staining method following the standard procedure described elsewhere8.
Identification of the lead resistant bacteria
Genomic DNA was isolated from the pure culture of the lead resistant bacteria and its quality was evaluated on 1.0% agarose gel. Fragment of 16S rDNA gene was amplified by 27F and 1492R primers. The PCR amplicon was purified to remove impurities. Forward and reverse DNA sequencing reaction of PCR amplicon was carried out with forward primer and reverse primers using BDT v3.1 Cycle sequencing kit on ABIxl Genetic Analyzer. Consensus sequence of 16Sr DNA gene was generated from forward and reverse sequence data using aligner software.
Sequence alignment and phylogenetic tree
The resultant gene sequence was used to carry out BLAST with the database of NCBI GenBank (https://blast.ncbi.nlm.nih.gov). Based on maximum identity score, first ten sequences were selected and aligned using multiple alignment software Clustal W. Distance matrix was generated and the phylogenetic tree was constructed using MEGA7 9. The evolutionary history was inferred by using the Maximum Likelihood method based on Kimura 2-parameter model 10. The bootsrap consensus tree inferred from 1000 replicates 11was taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pair-wise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with the superior log likelihood value. The analysis involved 11 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1434 positions in the final dataset.
Pot experiment to determine plant growth promoting properties
The healthy Indian mustard (Brassica juncea) and tomato (Solanum lycopersicum) seeds were collected from the local market. All the selected seeds were of uniform size. Garden soil was used to conduct the experiments. Soil samples were collected, air-dried, crushed, thoroughly mixed, and sieved to remove stones and debris. Seeds of B. juncea and S.lycopersicum were washed thoroughly with sterile de-ionized water. Then they were germinated in sterilized wet tissue paper. Four sets of pots were taken and filled with 500 g of collected soil and made wet with sterile distilled water. Fifty seeds of both species were sown in each pot. Four sets were prepared for both seeds: (i) Control (non-inoculated) (ii) Inoculated with lead resistant bacteria [T1] (106cfu/gm of soil) (iii) Inoculated with resistant bacteria and 1000ppm Pb(NO3)2 [T2] (iv) Soil with 1000ppm Pb(NO3)2 [T3] according to the method described 12 and modified to prepare the mentioned sets. All pots were allowed to grow for 4 weeks and were observed periodically for root, shoot and leaf development and results were recorded. The length taken from the above ground part of the sapling to the tip of the sapling was considered as the plant height. Average of that length is termed as average plant height. Length of the leaf was determined by measuring the length between leaf base and tip/apex of the leaf along midrib.
Statistical Analysis
All the experiments were done in triplicates. Data available from all the sets in pot experiments were treated with One-way ANOVA through SPSS software version 25.
Results
pH of the samples collected
Waste water samples were slightly alkaline in nature as evident from their pH values (Table 1).At sampling point 1, 6 and 8 pH of the collected water samples were close to 7.40 but sample 2 was little alkaline compare to them as pH at this point was 7.55. Highest alkalinity was observed in case of the water sample taken from sampling point 3having pH of 7.75. Wastewater samples from sampling points 4 and 5 were of intermediate alkalinity with pH of 7.60 and 7.65 respectively.
Table 1: pH of the collected wastewater samples.
|
Sampling point |
pH |
|
1 |
7.36 |
|
2 |
7.55 |
|
3 |
7.75 |
|
4 |
7.68 |
|
5 |
7.65 |
|
6 |
7.30 |
|
7 |
7.47 |
|
8 |
7.38 |
Enumeration of lead resistant bacteria from waste water samples
Amongst eight sampling points, number 3, 5 and 8 did not did not show any lead resistant bacteria. Rest water sources contained various numbers of lead resistant bacteria as mentioned in Table 2 in presence of 1000ppm of Pb(NO3)2. Maximum number of lead resistant bacteria i.e. 3430 colony forming units were observed from the waste water sample collected from sampling point 7 using 0.1ml. Wastewater sample from sampling point 1also showed presence of large number of lead resistant bacteria of more than 1700 in the same volume of water sample. Moderate number of colonies of 123 was calculated with the water sample from sampling point 2. Very few lead resistant colonies were isolated from sampling points 2 and 6.
Table 2: Colonies (Cfu/0.1ml) in water sample in presence of 1000 ppm of Pb(NO3)2
|
Sampling points |
1 |
2 |
4 |
6 |
7 |
|
No of colonies* |
1740 |
129 |
7 |
2 |
3430 |
Determination of minimum inhibitory concentration of lead towards the growth of the lead resistant bacteria
Lead resistant bacteria from five sampling points (1, 2, 4, 6 and 7) were grown in presence of higher concentrations of lead like 1200ppm, 1500ppm and 2000ppm to check the minimum inhibitory concentration of lead available for growth of the microbes. Table 3 showed that those resistant bacteria could tolerate 1000ppm of lead not higher. Lead resistant bacteria available from five sampling points showed observable growth in presence of 1000ppm of lead nitrate only. At a higher concentration of lead nitrate, the bacteria did not show observable growth which proved that growth of the lead tolerant bacteria present in the samples were inhibited in the presence of 1200ppm of lead nitrate and higher.
Table 3: Minimum inhibitory concentration of lead nitrate against the resistant bacteria
|
Metal concentration (ppm) |
1 |
2 |
4 |
6 |
7 |
|
1000 |
1740 |
129 |
7 |
2 |
3430 |
|
1200 |
– |
– |
– |
– |
– |
|
1500 |
– |
– |
– |
– |
– |
|
2000 |
– |
– |
– |
– |
– |
Table 4: Colony characteristics of the lead resistant bacteria available from five sampling points
|
Sampling Points |
Colony Characteristics |
||||||
|
Size |
Shape |
Texture |
Color |
Edge |
Elevation |
Opacity |
|
|
1 |
PP |
PF |
R |
CW |
W |
Flat |
Opaque |
|
2 |
M |
I |
R |
CW |
L |
Flat |
Opaque |
|
4 |
M |
C |
R |
CW |
I |
Raised |
Opaque |
|
6 |
PP |
F |
R |
BR |
E |
Flat |
Opaque |
|
7 |
PP |
PF |
R |
CW |
W |
Flat |
Opaque |
Size: PP– Pin point, M – Medium
Shape: PF – Puncti form, C – Circular, F – Filamentous, I – Irregular
Texture: R – Rough
Colour: CW – Creamy white, BR – Brown
Edge: E – Entire, W – Wavy, I – Irregular, L – Lobate
Colony characteristics of lead resistant bacteria
Lead resistant bacteria from five sampling points were grown in nutrient agar in presence of 1000ppm lead nitrate and colony characteristics of the resistant bacteria were noted as shown in Table 4. Colonies observed from the plated wastewater samples taken from sampling points 1, 2, 6 and 7 were rough in texture and flat in elevation. Amongst these, pin point sized, wavy edged and puncti form colonies were noted from the sampling points 1 and 7. Colonies from sampling point 2 were medium sized, irregular shaped and lobate edged whereas same from the sampling point 6 were pin point sized, filamentous and entire edged. Raised colonies were observed only in case of sampling point 4. These colonies were of medium size, circular, rough texture and irregular edge. All the colonies appeared as creamy white except for the sampling point 6, which were brown in colour.
Isolation of pure culture of lead resistant bacteria and identification of optimum pH and temperature
Among the lead resistant bacteria collected from five sampling points, bacteria from sampling point 7 were purified as single colony via spread plate method. All characterization experiments were done using the pure culture of the lead resistant bacteria. The resistant bacteria were gram positive in nature. Optimum pH and temperature for the growth of the lead resistant bacteria were experimentally determined as 7 and 50°C, respectively.
Antibiotic susceptibility test
Lead resistant bacteria were susceptible to several classes of antibiotics like penicillin, amoxyclav and ofloxacin as mentioned in Table 5.
Table 5: Antibiotic susceptibility of lead resistant bacteria
|
Serial Number |
Antibiotic used |
Concentration (mg/ml) |
Zone of inhibition (mm) |
Susceptible/Resistant |
|
1 |
Penicillin |
0.005 |
40 |
Susceptible |
|
0.010 |
44 |
Susceptible |
||
|
2 |
Amoxyclav |
0.005 |
41 |
Susceptible |
|
0.010 |
46 |
Susceptible |
||
|
3 |
Ofloxacin |
0.005 |
37 |
Susceptible |
|
0.010 |
50 |
Susceptible |
Identification of the lead resistant bacteria
A single band of high-molecular weight DNA had been observed when genomic DNA was resolved in 1% agarose gel (figure 1). During PCR amplification with forward and reverse primers, a single discrete PCR amplicon band of 1500bp was observed (Figure 2).
 |
Figure 1: Genomic DNA |
 |
Figure 2: 16 S PCR Amplicon. |
16S rDNA sequence was
5’-CCTCAGCGTCAGTTACAGACCAGAGAGTCGCCTTCGCCACTGGTGTTCCTCCACATCTCTACGCATTTCACCGCTACACGTGG AATTCCACTCTCCTCTTCTGCACTCAAGTTCCCCAGTTTCCAATGACCCTCCCCGGTTGAGCCGGGGGCTTTCACATCAGACTTA
AGAAACCGCCTGCGAGCCCTTTACGCCCAATAATTCCGGACAACGCTTGCCACCTACGTATTACCGCGGCTGCTGGCACGTAGT
TAGCCGTGGCTTTCTGGTTAGGTACCGTCAAGGTACCGCCCTATTCGAACGGTACTTGTTCTTCCCTAACAACAGAGCTTTACGA
TCCGAAAACCTTCATCACTCACGCGGCGTTGCTCCGTCAGACTTTCGTCCATTGCGGAAGATTCCCTACTGCTGCCTCCCGTAG
GAGTCTGGGCCGTGTCTCAGTCCCAGTGTGGCCGATCACCCTCTCAGGTCGGCTACGCAGTCGAGTTGCAGACTGCGATCCGA
ACTGAGAACAGATTTGTGGGATTGGCTTAACCTCGCGGTTTCGCTGCCCTTTGTTCTGTCCATTGTAGCACGTGTGTAGCCCAGG
TCATAAGGGGCATGATGATTTGACGTCATCCCCACCTTCCTCCGGTTTGTCACCGGCAGTCACCTTAGAGTGCCCAACTAAATGC
TGGCAAATAAGATCAAGGGTTGCGCTCGTTGCGGGACTTAACCCAACATCTCACGACACGAGCTGACGACACCCATGCCCCACCT
GTCACTCTGCCCCCGAAGGGGACGTCCTATCTCTAGGATTGTCAGAGGATGTCAAGACCTGGTAAGGTTCTTCG-3’.
Sequence alignment and Phylogenetic tree
Aligned query sequence was 99.79% identical with Bacillus subtilis strain IAM 12118 16S ribosomal RNA (Accession No. NR_112116.2) and others as given in Table 6.
Table 6: List of sequences producing significant alignments
|
Description |
Max |
Total |
Query |
E value |
Per.Ident |
Acc.Len |
Accession |
|
B. subtilis strain IAM 12118 16S ribosomal RNA, complete sequence |
887 |
1522 |
100% |
0 |
99.79% |
1550 |
NR_112116.2 |
|
B. subtilis subsp. inaquosorum strain BGSC 3A28 16S ribosomal RNA, partialsequence |
887 |
1527 |
100% |
0 |
99.79% |
1538 |
NR_104873.1 |
|
B. subtilis strain DSM 10 16S ribosomalRNA, partial sequence |
887 |
1527 |
100% |
0 |
99.79% |
1517 |
NR_027552.1 |
|
B. tequilensisstrain 10b 16S ribosomal RNA, partial sequence |
887 |
1527 |
100% |
0 |
99.79% |
1230 |
NR_117611.1 |
|
B. subtilis strain NRRL NRS-744 16S ribosomal RNA, partial sequence |
887 |
1479 |
97% |
0 |
99.79% |
1168 |
NR_116192.1 |
|
B. subtilis subsp. inaquosorum strain NRRL B-23052 16S ribosomal RNA, partialsequence |
887 |
1479 |
97% |
0 |
99.79% |
1168 |
NR_116188.1 |
|
B. subtilis subsp. spizizenii strain NRRL B-23049 16S ribosomal RNA, partial sequence |
887 |
1479 |
97% |
0 |
99.79% |
1168 |
NR_116187.1 |
|
B. subtilis strain NRRL B-4219 16S ribosomal RNA, partial sequence |
887 |
1479 |
97% |
0 |
99.79% |
1168 |
NR_116183.1 |
|
B. subtilis subsp. spizizenii strain NRRL B-23049 16S ribosomal RNA, partial sequence |
887 |
1527 |
100% |
0 |
99.79% |
1409 |
NR_024931.1 |
|
B. subtilis strain BCRC 10255 16S ribosomal RNA, partial sequence |
887 |
1522 |
100% |
0 |
99.79% |
1468 |
NR_116017.1 |
The evolutionary history was inferred by using the Maximum Likelihood method based on Kimura 2-parameter model 10. The bootsrap consensus tree inferred from 1000 replicates 11was taken to represent the evolutionary history of the taxa analyzed.
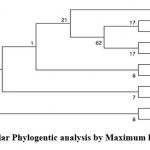 |
Figure 3: Molecular Phylogentic analysis by Maximum likelihood method. |
Pot experiment with the selected Bacillus subtilis strain
Germination of seeds was compared after two weeks as tomato seeds were germinating slowly (after 2 weeks) than mustard seeds (after one week). Both mustard and tomato seeds inoculated with B. subtilis germinated at a higher rate compared to the respective control samples consisting of only soil (Table 7). In case of mustard seeds 10percent greater germination was achieved in presence of B. subtilis but in case of tomato seeds it varied between ~7 to 13 percent.
Table 7: Germination percentage of seeds of Indian mustard & tomato (after 15 days)
|
Indian mustard (B. juncea) |
Tomato (S. lycopersicum) |
||||||
|
Control |
T1 |
T2 |
T3 |
Control |
T1 |
T2 |
T3 |
|
73.33 |
86.66 |
86.66 |
80.00 |
53.33 |
60.00 |
66.66 |
66.66 |
Control = Only soil
T1 = Soilwithculture
T2 = Soil with culture with 1000ppm Pb(NO3)2
T3 = Soilwith1000ppm Pb(NO3)2
In presence of 1000ppm Pb(NO3)2, growth of mustard plant was reduced as measured by the average height of the plant and leaf length after 7, 14, 21 and 28 days, respectively along with control set (Table 8).
Lead resistant B. subtilis augmented substrates showed improved average plant height and number of leaves than the mustard plants under metal stress (Table 8). After 7 days of growth, there was noted difference in average plant height between heavy metal laden B. subtilis supplemented and un-supplemented mustard saplings (Table 8). PGPR supplemented saplings achieved 2-3mm longer plants compared to the un-supplemented one. Similar augmentation was observed in other saplings where supplemented saplings were around 4mm longer. After 21days, supplemented saplings had grown 11mm longer plants than the uninoculated ones (Table 8). Similar trend was observed after 28days of growth also (Table 8). However, leaf numbers were not significantly different.
In case of leaf length, about 2mm longer leaves were noted in B. subtilis inoculated mustard saplings (Table 8) than un-inoculated plants after 7 days of growth. Comparable increment in leaf lengthwas spotted after 14days, 21 days and 28days of growth as well. Growth promoting activity of the inoculated Bacillus sp. was validated through statistical data (Table 9a and 9b). In each PGPR augmented mustard sapling (T1) higher average plant height and leaf length were detected after every week (Table 8) in comparison to all other sets. Evidently, the presence of the resistant bacteria encouraged the growth of the mustard plants under lead stress.
Table 8: Pot experiment with Indian mustard in presence of 1000ppm of Pb(NO3)2
|
Days Interval |
Average height (mm) |
Average no. of leaves |
Average leaf length (mm) |
|||||||||
|
Control |
T1 |
T2 |
T3 |
Control |
T1 |
T2 |
T3 |
Control |
T1 |
T2 |
T3 |
|
|
7 days |
14.53 |
18.27 |
16.07 |
14.47 |
2 |
2 |
2 |
2 |
7.40 |
10.20 |
8.67 |
7.53 |
|
14 days |
19.67 |
29.00 |
23.33 |
19.73 |
2 |
20 |
20 |
2 |
9.13 |
13.00 |
10.40 |
9.27 |
|
21 days |
30 |
56.87 |
39.67 |
30.07 |
2 |
2 |
2 |
2 |
11.33 |
15.2 |
13.67 |
11.4 |
|
28 days |
49.93a |
74.80b |
61.73a |
50.00a |
4 |
4 |
4 |
4 |
14.53p |
18.07q |
15.80r |
14.53p |
|
F test |
** |
NS |
** |
|||||||||
NS: non significant; *significant at p<0.05; ** significant at p<0.01
Mean values followed by same letter in each parameter do not differ significantly at P<0.05 using Tukey’s HSD (honestly significant difference).
There was significant difference of plant average height among the four pots in plant growth experiment p< 0.001(Table 8). For number of leaves, there was no significant difference between four pots in plant growth experiment p> 0.05 (Table 8).
In case of leaf length too, significant difference among the four pots was obtained p< 0.001(Table 8).
Most of the tomato saplings treated with 1000ppm Pb(NO3)2 showed measurable differences of around 1mm and 2mm in leaf length till 21days and 28days of growth respectively in presence of PGPR.
Table 9: Pot experiment with tomato in presence of 1000ppm of Pb(NO3)2
|
Days Interval |
Average Height (mm) |
Number of leaf |
Leaf length (mm) |
|||||||||
|
Control |
T1 |
T2 |
T3 |
Control |
T1 |
T2 |
T3 |
Control |
T1 |
T2 |
T3 |
|
|
14 days |
12.73 |
15.67 |
14.53 |
12.93 |
2 |
2 |
2 |
2 |
4.13 |
5.60 |
5.20 |
4.27 |
|
21 days |
17.73 |
22.07 |
18.13 |
17.80 |
2 |
2 |
2 |
2 |
7.80 |
8.93 |
7.73 |
7.87 |
|
28 days |
52.53 |
70.00 |
60.20 |
52.67 |
4 |
4 |
4 |
4 |
14.07 |
18.13 |
16.07 |
14.07 |
|
F test |
NS |
NS |
NS |
|||||||||
NS: non significant
But, analyzed data obtained through one-way ANOVA showed that those differences among the four pots in plant growth experiment were not significant as all the sets were more or less similar with respect to plant height, number of leaves and leaf length (Table 9). Although, presence of B. subtilis in heavy metal laden soil resulted in notable improvement in average plant height and leaf length of Indian mustard saplings but similar achievement was unattainable in case of tomato saplings (Table 8 and 9). Moreover, B. Subtilis augmented germination of both the seeds albeit at different rates (Table 7).
Discussion
Bacillus subtilis is amongst the common plant growth promoting rhizobacteria (PGPR), which is abundant in soil ecosystem and plays significant role in mitigating biotic and abiotic stresses faced by plant species. These bacteria confer stress tolerance to the plants by several mechanisms like induced systemic resistance, biofilm formation and lipopeptide production etc. to name a few5. It also acts as a potent denitrifying agent as well. PGPRs directly interact with the plant species and help uptake of both macro and micronutrients, production of phytohormones, regulate the release of stress hormones and enzymes like ethylene, abscisic acid and 1-aminocyclopropane 1-carboxylase deaminase etc. These bacteria have a passive role as biocontrol agent by the virtue of their cell wall lysing enzymes and lipopeptide production etc. Sun et al, 202013 reported of Bacillus sp. as inoculants in farmlands to reduce the release of nitrogen and ammonia gases. It also critically regulates biogeochemical cycles and provides proper aeration to the soil ecosystem. Bacillus subtilis usually provide intrinsic resistance against a broad spectrum of antibiotics because of the presence of membrane efflux pumps and BceAB type transporters in them14. Lead resistant B. subtilis isolated from industrial waste water (from sampling point 7) showed plant growth promoting activity on Indian mustard in presence of 1000ppm of Pb(NO3)2 but in case of tomato plants similar trend was not observed. However, those PGPR promoted germination of both the seeds in presence of heavy metal. Slow germination rate in case of tomato seeds may be attributed to different growth requirements of the respective plant compare to Indian mustard under the experimental condition. Moreover, it might be possible that applied PGPR inoculum was insufficient for the stressed tomato plants to alleviate metal stress.
Bacillus subtilis FBL-10 mitigated lead toxicity in Solanum melongena via activation of antioxidant system of the plant and at the same time decreased the level of peroxide and malonaldehyde15. In Chick pea, B. subtilis increased proline content and decreased lipid peroxidation during nickel, lead and cadmium metal stress16. Bacillus subtilis PBRB3 encouraged higher antioxidant enzyme activity in Mung bean under lead stress17. Similar toxicity induced regulation of homeostasis of anti-oxidants in radish in presence of B. subtilis CIK512 18. Bacillus subtilis BM2 improved overall growth parameters of wheat grown under metal stress. In presence of 195mg Pb/Kg, it enhanced the length and dry biomass of shoots by 14% and 23%, respectively over the control19. The strain also improved grain yield significantly by 55% at 585mg Pb/Kg compared to uninoculated plants. Bacillus subtilis BM2 relieved the metal stress on wheat by lowering the levels of proline and malonaldehyde along with the activities of antioxidant enzymes like catalase, superoxide dismutase and glutathione reductase etc. Bacillus subtilis KW1 isolated from rhizosperic soil promoted growth of Chinese cabbage and rice cultivars through secretion of bioactive compounds20. Presence of B. subtilis resulted in significant increase in plant height, biomass, chlorophyll contents and nutrient uptake.
In the present study, presence of lead resistant B. subtilis isolated from petrochemical waste water ameliorated lead stress in Indian mustard saplings and promoted growth of the saplings with increased average plant height and leaf length. Incidentally B. subtilis isolated from the industrial waste waters were susceptible to various antibiotics (Table 5) indicating some alterations in the genetic makeup of the lead resistant strain. This observation can be explored further through genomic, transcriptomic, proteomic or metabolomic studies to assess structural and functional parameters of the concerned genes and related products available inthe isolated strain. Antibiotic resistant genes of PGPRs often pose threat to the ecosystem as they sometimes affect the endophytic and rhizospheric microbiota via horizontal gene transfer. But, the isolated lead resistant B. subtilis is devoid of such concern and can be applied as ecofriendly biofertilizer to the fields after successful field trials. These bacteria can be applied to other heavy metal laden soils to check their PGPR activity in future and will be greatly beneficial in the management of metal polluted soils.
Acknowledgement
The authors are deeply indebted to Dr. Jayasree Laha, Principal, Raja N. L. Khan Women’s College, Paschim Medinipur and Dr. Sasabindu Jana, Principal, Raidighi College, South 24 Parganas. Sincere acknowledgement to DST-Curie for infrastructural support to Natural and Applied Sciences Research Centre, Raja N. L. Khan Women’s College, Paschim Medinipur.
Conflict of Interest
All the authors declare that there is no conflict of interest with any authority.
Funding source
The authors declare that the expenditure incurred during the experiments was contributed by the corresponding author’s institutional authorities. No external fund was utilized in the process.
References
- Yan A., Wang Y., Tan S. N., Mohd Yusof M. L., Ghosh S.,Chen Z. Phytoremediation: A Promising Approach for Revegetation of Heavy Metal-Polluted Land. Front Plant Sci. 2020; 11:359.
CrossRef - Ma Y., Prasad M. N. V., Rajkumar M., Freitas H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol Adv. 2011; 29(2): 248–258.
CrossRef - Vamerali T., Bandiera M., Mosca G. Field crops for phytoremediation of metal-contaminated land. A review. Environ Chem Lett. 2010; 8: 1–17.
CrossRef - DalCorso G., Fasani E., Manara A., Visioli G., Furini A. Heavy metal pollutions: state of the art and innovation in phytoremediation. Int J Mol Sci. 2019; 20(14):3412.
CrossRef - Mahapatra S., Yadav R., Ramakrishna W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J ApplMicrobiol. 2022; 132(5):3543–3562.
CrossRef - Yamina B., Tahar B., Marie-Laure F. Isolation and screening of heavy metal resistant bacteria from wastewater: a study of heavy metal co-resistance and antibiotic resistance. Water Sci Technol. 2012; 66(10), 2041-2048.
CrossRef - Gupta K., Chatterjee C., Gupta B. Isolation and characterization of heavy metal tolerant Gram-positive bacteria with bioremedial properties from municipal waste rich soil of Kestopur canal (Kolkata), West Bengal. Biologia. 67(5): 827-836.
CrossRef - Bartholomew J. W., Mittwer T. The Gram stain. Bacteriol Rev.1952; 16 (1) 1-29.
CrossRef - Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol BiolEvol. 2016; 33(7), 1870-1874.
CrossRef - Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111-120.
CrossRef - Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985; 39(4):783-791.
CrossRef - Seher N-u., Ahmad M., Ahmad I., Nazli F., Mumtaz M. Z., Latif M., Al-Mosallam M. S., Alotaibi F. S., Dewidar A. Z., Matter M. A., El-Shafei A. A. Lead-tolerant Bacillus strains promote growth and antioxidant activities of spinach (Spinaciaoleracea) treated with sewage water. Agronomy. 2021; 11: 2482.
CrossRef - Sun B., Bai Z., Bao L., Xue L., Zhang S., Wei Y., Zhang Z., Zhuang G., Zhuang X. Bacillus subtilisbiofertilizer mitigating agricultural ammonia emission and shifting soil nitrogen cycling microbiomes. Environ Int. 2020; 144: 105989.
CrossRef - Rismondo J., Schulz, L. M. Not Just Transporters: Alternative functions of ABC transporters in Bacillus subtilis and Listeria monocytogenes. Microorganisms. 2021; 9(1): 163.
CrossRef - Shah A., Yasin N.A., Akram K., Ahmad A., Khan W.U., Akram W., Akbar M. Ameliorative role of Bacillus subtilis FBL-10 and silicon against lead induced stress in Solanummelongena. Plant PhysiolBiochem. 2021; 158: 486-496.
CrossRef - Khan N., Bano, A. Effects of exogenously applied salicylic acid and putrescine alone and in combination with rhizobacteria on the phytoremediation of heavy metals and chickpea growth in sandy soil. Int J Phytoremediation.2018; 20(5): 405-414.
CrossRef - Arif M.S., Yasmeen T., Shahzad S.M., Riaz M., Rizwan, M., Iqbal S., Asif M., Soliman M. H., Ali S. Lead toxicity induced phytotoxic effects on mung bean can be relegated by lead tolerant Bacillus subtilis (PbRB3). Chemosphere. 2019; 234: 70-80.
CrossRef - Ahmad I., Akhtar M.J., Mehmood S., Akhter K., Tahir M., Saeed M.F., Hussain M. B., Hussain S. Combined application of compost and Bacillus sp. CIK-512 ameliorated the lead toxicity in radish by regulating the homeostasis of antioxidants and lead. Ecotoxicol Environ Saf. 2018; 148: 805-812.
CrossRef - Rizvi A., Ahmed B., Zidi A., Khan M. S. Heavy metal mediated phytotoxic impact on winter wheat: oxidative stress and microbial management of toxicity by Bacillus subtilis RSC Adv. 2019; 9(11): 6125-6142.
CrossRef - Kang S-M., Hamayun M., Khan M. A., Iqbal A., Lee I-J. Bacillus subtilis JW1 enhances plant growth and nutrient uptake of Chinese cabbage through gibberellins secretion. J Appl Bot Food Qual. 2019; 92(1): 172-178.

