Huanglongbing (HLB) is the most serious disease of citrus, putatively caused by the bacterium ‘Candidatus Liberibacter asiaticus’ (CLas), transmitted by the Asian citrus psyllid (Diaphorina citri Kuwayama) in the USA. Most commercial citrus cultivars are susceptible to the disease, particularly conventional sweet oranges and grapefruit, decline severely in health and productivity (Bové 2006, Gottwald 2010). Early detection and prompt response are key factors in the eradication or suppression of HLB epidemics in California. Real-time PCR is the standard regulatory method for CLas detection and it can detect as little as a single copy of bacterium in a sample. However, due to lack of visible symptoms, low titer and uneven distribution of CLas in a tree, selecting the best sample is a major hurdle for early detection by qPCR. Regulatory testing of CLas by qPCR stipulates sampling of mature leaves. Such leaves are good for visual inspection of HLB symptoms but can be unreliable tissue for early detection and low titer infections. Symptoms of HLB develop slowly and without symptoms, collecting that right leaf/shoot amongst >200,000 estimated leaves per mature tree is practically impossible for early diagnosis.
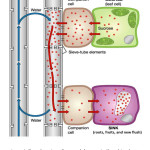 |
Figure 1: Translocation of sugars (photoassimilates) in plants. Pressure flow hypothesis/ Münch mechanism by which sugars are transported through the phloem, from sources to sinks tissues. Click here to View Figure |
The pressure flow hypothesis or Münch mechanism states that sugars, made in the leaves via photosynthesis, move passively down a concentration gradient, or actively transported in some cases (Fig 1). Accumulation of sugar (photoassimilates) in phloem osmotically attracts water, increasing the pressure in the sieve tubes at the source. This pressure pushes phloem sap towards a region of lower pressure, consequently, the sap moves by bulk flow, down a pressure gradient (Jensen et al., 2016). Phloem-limited plant pathogens move in a source-to-sink fashion along with photoassimilates (Bendix and Lewis, 2018). New flush has been the reliable tissue to test Citrus tristeza virus (CTV) and Citrus Pest Detection Program (CPDP) of Central California Tristeza Eradication Agency has been utilizing the flush tissue for large-scale field survey for the last three decades or so (Gottwald and Hughes, 2000). Recent findings indicate new flush can be used for reliable detection of CLas (McCollum, unpublished).
Bar-Joseph et al., (1979) found higher content of CTV in peduncle bark than the bark of branches of same age. Similarly, our preliminary data of testing Spiroplasma citri from young shoot indicates that the titer of this phloem-limited bacteria is more in peduncle of young fruit than the bark of young shoot and petiole of young leaf (Hajeri and Yokomi, unpublished). Moreover, fruit tissues such as columella and receptacle are routinely used for reliable detection of S. citri by CPDP (Yokomi et al., 2008). In Florida, Tatineni et al., (2008) and in Texas, Kunta et al., (2014) observed a relatively high titer of CLas in fruit peduncle. Recently, in Texas, Park et al (2018) have shown that the CLas can be detected in roots before appearance of visible symptoms aboveground. Moreover, CLas titer was not only greater in the roots but also uniformly distributed, and more consistent through the year when compared to leaf samples (Johnson et al., 2014, Louzada et al., 2016, and Park et al., 2018). In all the cases, the sink tissue such as the new flush, fruit and roots showed high titer of phloem-limited citrus pathogens. In Florida, Irey et al. (2011) observed that the greatest percentage of HLB positives by qPCR were during July through January, coinciding with the period of maximum leaf symptomatology and concluded that seasonality of sampling is critical.
The outbreak of HLB in California is delimited to residential properties in four southern California counties: Los Angeles, Orange, Riverside and San Bernardino; whereas, no detection of HLB has occurred in any commercial citrus orchard in California. Rapid detection and eradication of infected trees is the highest priority along with monitoring and control of D. citri, vector of CLas in commercial orchards of California.
Conclusion
Early detection of CLas, long before symptom appearance, is a critical need in California. Moreover, regulatory detection currently requires direct pathogen detection and quantitation of target DNA of CLas. When it comes to sampling large trees in commercial setting, data gap exists despite valuable data available from previous studies. There is a need for systematic sampling study to identify the most reliable tissue matched with its ideal season for early detection of CLas in mature citrus trees.
References
- Bar-Joseph M, Garnsey SM, Gonsalves D, Moscovitz M, Purcifull DE, Clark MF and Loebenstein G. 1979. The use of enzyme-linked immunosorbent assay for detection of Citrus tristeza virus. Phytopathol. 69(2):190-194.
- Bendix C and Lewis JD. 2018. The enemy within: phloem-limited pathogens. Mol. Plant Pathol. 19:238-254.
- Bove JM. 2006. Huanglongbing: a destructive, newly emerging, century-old disease of citrus. J Plant Pathol. 88, 7–37.
- Gottwald TR and Hughes G. 2000. A new survey method for citrus tristeza virus disease assessment. In: Proceedings of the 14th Conference of the International Organization of Citrus Virologists, 77–87.
- Gottwald, T. R. 2010. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48: 119–139.
- Irey, MS, Gast T, Cote J, Gadea P, Santiago O, Briefman L, and Graham, JH. 2011. Seasonal variability in HLB testing data in plant and psyllid samples in Florida. Page 72 in: Proc. 2nd Int. Res. Conf. Huanglongbing, Orlando, FL.
- Jensen KH, Berg-Sorensen K, Bruus H, Holbrook NM, Liesche J, Schulz A, Zwieniecki MA, and Bohr T. 2016. Sap flow and sugar transport in plants. Rev. Mod. Phys. 88:035007.
- Johnson EG, Wu J, Bright DB, and Graham JH. 2014. Association of ‘Candidatus Liberibacter asiaticus’ root infection, but not phloem plugging with root loss on huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathol. 63:290-298.
- Kunta M, da Graca JV, Malik NSA, Louzada E, and M. Setamou. 2014. Quantitative distribution of Candidatus Liberibacter asiaticus in the aerial parts of the Huanglongbing-infected citrus trees in Texas. HortScience 49(1):65-68.
- Louzada ES, Vazquez OE, Braswell WE, Yanev G, Devanaboina M, and Kunta M. 2016. Distribution of ‘Candidatus Liberibacter asiaticus’ above and below ground in Texas citrus. Phytopathol. 106: 1-8.
- Park JW, Louzada ES, Braswell WE, Stansly PA, da Graça JV, McCollum G, Rascoe JE and Kunta M. 2018. A new diagnostic real-time PCR method for huanglongbing detection in citrus root tissue. J Gen. Pl. Pathol. 84(5):359-367.
- Tatineni S, Sagaram US, Gowda S, CJ Robertson, CJ, Dawson WO, Iwanami T and Wang N. 2008. In plant distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathol. 98:592–599.
- Yokomi RK, Mello AF, Saponari M and Fletcher J. 2008. Polymerase chain reaction-based detection of Spiroplasma citri associated with citrus stubborn disease. Pl. Dis. 92(2):253-260.

