Introduction
Increasing plant production and the quality of nutrients in the food is a major concern now a days. The plant production is affected by many factors such as climate conditions, rising temperatures, untimely flood, lack of moisture in the air, increase/decrease in the levels of CO2 and nitrogen, pests and pathogens etc. The plants get affected by these factors and catch diseases due to bacterial, fungal infections which can spread quickly. The symptoms of the disease can occur on stems, leaves and fruits of the plant. It is of great concern to take preventive action immediately else it can spoil the plants and affect the plant productivity. Technology can help the farmers take the right decision about the use of appropriate methods/pesticides to eradicate or control the spread of the disease.
Deep learning models have been widely experimented with large domains such as computer vision, NLP etc. and their success is demonstrated in various related applications. The PDD researchers are also employing deep learning models for accurately detecting the diseases. The major issue that the PDD researchers face is the limited set of data. Deeper and deeper networks with high capacity are being built but due to lack of sufficient data these models can give over optimistic results. Hence, it is necessary to train the deep learning models with adequate data. Image Data Augmentation Techniques (IDA) can come to the rescue to address this scarcity. IDA is the process of generating synthetic images to increase the dataset size. IDA involves applying some transformation operation on the image that can generate a new image where the relevant features/structures are retained, and irrelevant features/structures are modified.
In the literature1-17, several authors have discussed importance of IDA for deep learning based applications. Many IDA techniques have been developed and the most recent ones involve latent space-based techniques using Generative Adversarial Networks (GAN)4,7 . The GAN can generate images using a model consisting of generator and discriminator. These models are trained together in an adversarial manner where the generator generates fake images from a random image. The discriminator takes the images generated by the generator and recognizes it as fake/real. The GAN can generate a huge number of realistic images but are computationally complex.
The contribution of the research proposed in this article is as follows. First, we have developed two mixing methods for IDA. Hadamard transform and traditional transforms such as rotation and flipping are used for developing mixing methods. The mixing methods use the feature space (frequency domain coefficients) obtained after applying the Hadamard transform on two images. These coefficients are mixed carefully after following the theoretical analysis to generate images. Since IDA is also applied to address the issue of generalization, this study also compares the results of the proposed mixing methods to regularization techniques such as drop out (DO)14,18 Batch normalization (BN)14,18 and their combinations that are applied to address the issue of generalization. The proposed study is carried out on the Plant Village Dataset and three convolutional neural networks (CNN) VGG1619, VGG1919 ResNET-5019.
Literature Review
In this section, the review of PDD using deep learning-based models and a few IDA techniques is presented.
Recently leveraging deep learning technologies have become the state-of-the-art methods for PDD1-11. Colour Manipulation methods such as brightness, contrast, sharpness etc. are studied for detecting single disease in horticulture plants using region based fully connected network and found that these techniques are effective only or certain categories2. The effectiveness of EfficientNetV2 for detecting two diseases in Cardamom plants and three diseases in grapes is confirmed by the experimental study3. Six different CNN models are trained for detecting severity in citrus diseases and Deep Convolutional Generative Adversarial Networks (DCGAN) based DA method is proposed to augment the dataset. This research specified that the GAN based IDA method is effective and improved the accuracy of InceptionV3 model4. Lightweight MInception with MobileNet is proposed to extract better features for PDD6. DoubleGAN is applied to generate images of unhealthy leaves to address the issue of unbalanced dataset7. This model used healthy images in stage 1 to pretrain the WGAN model and in stage 2 to generate images of size 64X64 using unhealthy leaves images. A deep learning framework using EfficientNet-13 and Bidirectional feature fusion (BiFN) is proposed to classify diseased images and locate infected portions of the disease leaves8. EfficientNet was applied to learn finer characteristics and BiFN for feature fusion. A class and box prediction network predicted the class of plant leaves and generated box coordinates to identify infected images. Better performance is observed with transfer learning and DA than state-of-the-art methods8. A CNN based classifier is applied for tomato disease detection, with a performance above 95% accuracy11. IDA methods are categorized into manual methods and automated DA methods12. In manual methods applying traditional methods such as translation, rotation, shear, erase, swirl, horizontal flip, dilation, erosion, colour manipulation methods etc is common. In automated DA (AutoDA), DA policies are selected automatically. AutoDA methods are categorized into composition, mixing and generation-based approaches. The composition-based methods apply techniques such as reinforcement learning, Bayesian network, Gradient Descent etc. to search for the best choice and parameters of operations. The mixing methods create mixed images by applying some mixing strategy for example neural mixing uses neural network and multiple images to create a mixed image. Generation based IDA apply GAN based methods to synthesize new images. Style transfer techniques generate new images by transferring the style of one image onto the other retaining the important features12. Generative models based diffusion model was applied to generate synthetic images and augment the datasets with diverse images13. Authors applied Fréchet Inception Distance (FID) and Kernel Inception Distance (KID) to demonstrate the quality of the images generated.
Shorten and Khoshgoftaar14 presented a comprehensive survey of IDA techniques. This article discussed various IDA methods, and the review suggestd that the feature space augmentations are less attempted. Research carried out by Hernández-García and König15, concluded that the DA methods are more adaptable to changes in the parameters such as network architecture compared to regularization methods such as weight decay and drop out. Few researchers have attempted frequency domain-based IDA16,17 where orthogonal transforms were used to obtain the feature space. The images were generated by manipulating and then transforming this feature space.
To summarize, research in IDA methods is advanced and integral part of performance improvement in image classification tasks including PDD.
Materials and Methods
The image mixing methods proposed in this research are applied for plant disease detection. Geometric Transformations(GT) such as horizontal flipping and rotation=180 are applied on the original image to obtain GTimage. Proposed mixing methods use the original image and GTimage to create a new image and enhance the dataset. The general strategy of the proposed approach is given in Figure 1 and described below. .
Stage-1 Obtain the Coefficient Matrix (CM)
The Red,Green, Blue colour channels of the image and the GTimage are obtained.
Hadamard matrix16 is applied on each of the six channels using and six Coefficient Matrices (CM) (TR-1, TG-1,TB-1,TR-2,TG-2,TB-2) are generated as shown in Figure 1. These 6 CMs are used for mixing in stage 2.
Stage-2 Mixing the Coefficients of CM
The CMs represent the frequency domain components of the respective channels. The coefficients related to the most relevant features/high frequency components of the channels are concentrated in the top-left corner of the matrix. The least relevant features or the low frequency components that have less information are concentrated in the bottom right corner of the matrix. Here we mix the low frequency components of two images to generate a new image. The following approaches have been applied in this work.
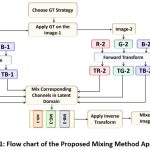 |
Figure 1: Flow chart of the Proposed Mixing Method Approach |
Mixing Method 1
Here, the CMs for the input image and flipped input image are mixed to obtain the new CM. Average of the lower frequncy coeffcients are taken to generate the mixed images.
Mixing Method 2
Here, the CMs for the input image and input image rotated by 180 are mixed to obtain the new CM. The mixing strategy uses the frequency components in the ratio of (2/3 input image transformed +1/3 GTimage transformed).
Stage-3 Generating Synthetic Images
The CMs obtained after mixing procedure in Stage-2 are used for synthesis of new images. Inverse Hadamard transform16 is applied on the CM to obtain new images.
PDD Methodology
The methodology used for Plant Disese Detection is shown in Figure 2 and briefly described here. For PDD, the proposed IDA technique as described in Figure 1 is applied on the training set to increase the dataset size. The datset is augmented with the proposed mixing methods. The VGG1619, VGG1919 and ResNet5020 models are trained on the augmented dataset while testing set is kept unaugmented. The selected model is trained with the given hyper-parmeters using different experiments viz. training with the orginal dataset, training with augmented dataset, training with functional regularization techniques such as Drop out14,18 (DO), Batch Normalization14,18(BN) and their combinations and traditional data augmentations (DA). Performance evaluation is done using the metrics as given below in equation (1) to equation (4).
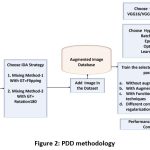 |
Figure 2: PDD Methodology |
Performance Evaluation
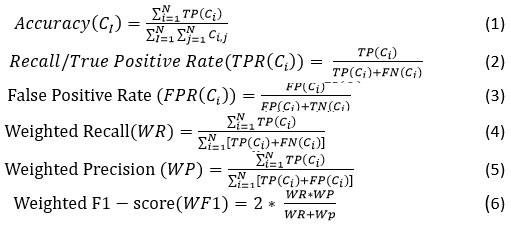
Performance evaluation metrics21 as given in Eq. (1)-Eq. (6) are applied. Here, TP: True Positive, FP: False Positive, TN: True Negative, FN: False Negative, Ci: ith Class.
PlantVillage Dataset
PlantVillage dataset22,23 is created by Hughes and Salathe in 2016 through an online platform using crowdsourcing. The dataset contains around 54305 colour images of leaves from 14 different plant species. This benchmark dataset contains healthy and disease leaves images labelled by experts. The images are split into 32571 training and 21734 testing images.
Results and Discussion
Octave environment is used for image processing part. Google Colab is used with GPU settings for the PDD using VGG16, VGG19 and ResNet50 models. The models are trained for 125 epochs, batch size=64, adam optimizer and learning rate=0.001. Performance evaluation is done using accuracy, precision, recall and F1-score. Results of the proposed mixing methods are compared with following DA method: zoom = 0.2, rotation = 30, width_shift = 0.2, height_shift = 0.2, shear = 0.2. Comparison of the proposed mixing methods is also done with drop out (DO) with DO =0.4 and batch normalization (BN).
Results Analysis
Performance of the results obtained for VGG16, VGG19 and ResNet50 are presented in Tables 1-3. The graphs of accuracies are presented in Figure 3.
Table 1: Performance evaluation of the results obtained for VGG-16
|
Method |
Accuracy |
Precision |
Recall |
F1- S |
|
No Aug/Reg. |
0.8913 |
0.8914 |
0.8913 |
0.8910 |
|
Drop Out |
0.9028 |
0.9024 |
0.9028 |
0.9016 |
|
BN |
0.8761 |
0.8766 |
0.8761 |
0.7744 |
|
DA |
0.7418 |
0.7607 |
0.7418 |
0.7327 |
|
BN+DO |
0.8725 |
0.8743 |
0.8725 |
0.8724 |
|
DA+BN+DO |
0.3344 |
0.5408 |
0.3344 |
0.3503 |
|
Mixing- 1 |
0.8261 |
0.8289 |
0.8261 |
0.8264 |
|
Mixing -2 |
0.8216 |
0.8224 |
0.8216 |
0.8217 |
Table 2: Performance evaluation of the results obtained for VGG-19
|
Method |
Accuracy |
Precision |
Recall |
F1- S |
|
No Aug/Reg |
0.8691 |
0.8705 |
0.8691 |
0.8691 |
|
Drop Out |
0.8830 |
0.8834 |
0.8830 |
0.8813 |
|
BN |
0.8542 |
0.8602 |
0.8542 |
0.8552 |
|
DA |
0.7623 |
0.7784 |
0.7623 |
0.7516 |
|
BN+DO |
0.8719 |
0.8712 |
0.8719 |
0.8706 |
|
DA+BN+DO |
0.6211 |
0.6114 |
0.6211 |
0.5897 |
|
Mixing- 1 |
0.8213 |
0.8249 |
0.8213 |
0.8220 |
|
Mixing -2 |
0.8037 |
0.8080 |
0.8037 |
0.8034 |
Accuracy of VGG16 is better than VGG19 and ResNet50 when experiment is carried out on the original dataset. Drop-out (DO) worked very well for VGG16 followed by VGG19, however it has average performance on ResNet50. For VGG19, a combination of BN+DO has better performance than other methods. The performance of the proposed mixing methods has significantly improved for ResNet50, whereas for VGG16 and VGG19 the performance is average. This analysis confirms that all the regularization techniques cannot be applied uniformly to all the selected models.
Table 3: Performance evaluation of the results obtained for ResNet50
|
Method |
Accuracy |
Precision |
Recall |
F1- S |
|
No Aug/Reg |
0.7024 |
0.7336 |
0.7024 |
0.6995 |
|
Drop Out |
0.5920 |
0.5870 |
0.5920 |
0.5538 |
|
BN |
0.6862 |
0.6974 |
0.6862 |
0.6848 |
|
DA |
0.1897 |
0.0661 |
0.1897 |
0.0898 |
|
BN+DO |
0.6469 |
0.6310 |
0.6469 |
0.6211 |
|
DA+BN+DO |
0.1988 |
0.1172 |
0.1988 |
0.0952 |
|
Mixing- 1 |
0.9426 |
0.9438 |
0.9426 |
0.9421 |
|
Mixing -2 |
0.9432 |
0.9445 |
0.9432 |
0.9430 |
The comparison graph of the accuracies is presented in Figure 3 as shown below.
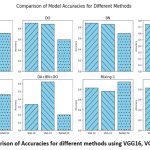 |
Figure 3: Comparison of Accuracies for different methods using VGG16, VGG19, ResNet50. |
The pros and cons of the proposed mixing methods are briefly discussed here. The proposed methods can easily generate new images just by varying number of coefficients (high frequency component and low frequency components) while mixing. This introduces some diversity in the images. Huge number of images can be generated in short span of time without training complex networks such as GAN. However, the quality of the image generated after mixing depends on the two images. The resultant image may show slight blur and hazy effect. Two different images of the same object with completely different background can generate an image that might look unrealistic. Further, the images used for mixing must be of the same size and from same distribution.
Conclusion
This research leveraged latent space obtained from Hadamard transform and proposed two mixing methods for IDA. The proposed methods utilized the latent spaces of original images and geometrically transformed images for generating new images. The performance of the methods is evaluated using VGG16, VGG19 and ResNet50 for plant disease detection task using PlantVillage dataset. The proposed methods are simple, easy to implement, do not require complex models for training and are promising for generating images with diversity. The proposed methods have shown significant improvement in performance for ResNet50 model. Overall mixing method 2 has performed better than mixing method 1. Comparison of the proposed methods with traditional DA methods revealed significant improvement in performance for all the three models.
Acknowledgement
The authors are thankful to the Principal, Thadomal Shahani Engineering College, Mumbai for providing the necessary resources for carrying out this research.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
PlantVillage Dataset isavailable at: https://data.mendeley.com/datasets/tywbtsjrjv/1. Accessed August 22, 2024.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Author contributions
Vaishali Suryawanshi: The proposed IDA mixing method was conceptualized, implemented and articulated in this article
Sahil Adivarekar: Supporting results were carried out
Tanuja Sarode: Reviewed the whole process including the research article.
References
- Li L, Zhang S, Wang B. Plant disease detection and classification by deep learning—a review. IEEE Access. 2021;9:56683-56698. doi:10.1109/ACCESS.2021.3069646.
CrossRef - Saleem M. H., Potgieter J, Arif K. M. A performance-optimized deep learning-based plant disease detection approach for horticultural crops of New Zealand. IEEE Access. 2022;10:89798-89822. doi:10.1109/ACCESS.2022.3201104.
CrossRef - Kumar C., Jaidhar C. D., Patil N. Cardamom plant disease detection approach using EfficientNetV2. IEEE Access. 2022;10:789-804. doi:10.1109/ACCESS.2021.3138920.
CrossRef - Zeng Q, Ma X, Cheng B, Zhou E, Pang W. GANs-based data augmentation for citrus disease severity detection using deep learning. IEEE Access. 2020;8:172882-172891. doi:10.1109/ACCESS.2020. 3025196.
CrossRef - Tabbakh A, Barpanda SS. A deep features extraction model based on the transfer learning model and vision transformer “TLMViT” for plant disease classification. IEEE Access. 2023;11:45377-45392. doi:10.1109/ACCESS.2023.3273317.
CrossRef - Chen J, Chen W, Zeb A, Yang S, Zhang D. Lightweight inception networks for the recognition and detection of rice plant diseases. IEEE Sens J. 2022;22(14):14628-14638. doi:10.1109/ JSEN.2022. 3182304.
CrossRef
- Zhao Y, Chen Z, Gao X, Song W, Xiong Q, Hu J. Plant disease detection using generated leaves based on DoubleGAN. IEEE/ACM Trans Comput Biol Bioinformatics. 2022;19(3):1817-1826. doi:10.1109/TCBB.2021.3056683.
CrossRef - Lakshmi RK, Savarimuthu N. PLDD—a deep learning-based plant leaf disease detection. IEEE Consum Electron Mag. 2022;11(3):44-49. doi:10.1109/MCE.2021.3083976.
CrossRef - Liu Z, Bashir RN, Iqbal S, Shahid MMA, Tausif M, Umer Q. Internet of things (IoT) and machine learning model of plant disease prediction–blister blight for tea plant. IEEE Access. 2022;10:44934-44944. doi:10.1109/ACCESS.2022.3169147.
CrossRef - Delnevo G, Girau R, Ceccarini C, Prandi C. A deep learning and social IoT approach for plants disease prediction toward a sustainable agriculture. IEEE Internet Things J. 2022;9(10):7243-7250. doi:10.1109/JIOT.2021.3097379.
CrossRef - Kokate JK, Kumar S, Kulkarni A. G. Classification of Tomato Leaf Disease Using a Custom Convolutional Neural Network. Curr Agri Res 2023; 11(1). doi : http://dx.doi.org/10.12944/ CARJ.11.1.28.
CrossRef - Yang Z, Sinnott RO, Bailey J, Ke Q. A survey of automated data augmentation algorithms for deep learning-based image classification tasks. Knowl Inf Syst. 2023;65(7):2805-2861. https://doi.org/ 10.1007/s10115-023-01853-2.
CrossRef - Muhammad A, Salman Z, Lee K, Han D. Harnessing the power of diffusion models for plant disease image augmentation. Front Plant Sci. 2023 Nov 7;14:1280496. https://doi.org/10.3389/ fpls.2023.1280496.
CrossRef - Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1). https://doi.org/10.1186/s40537-019-0197-0.
CrossRef - Hernández-García A, König P. Further advantages of data augmentation on convolutional neural networks. Paper presented at: International Conference on Artificial Neural Networks (ICANN); October 4-7, 2018; Rhodes (Greece). https://doi.org/10.1007/978-3-030-01418-6_10.
CrossRef - Suryawanshi V, Sarode T, Jhunjhunwala N, Khan H. Evaluating image data augmentation technique utilizing Hadamard Walsh space for image classification. Paper presented at: International Conference on Intelligent Vision and Computing (ICIVC); November 26-27, 2022; Agartala. https://doi.org/10.1007/978-3-031-31164-2_24. Accessed 23 July 2024.
CrossRef - Suryawanshi V, Sarode T. Performance analysis of DCT-based latent space image data augmentation technique. Paper presented at: 6th International Conference on Innovative Computing and Communications (ICICC); February 17-18, 2023; New Delhi. DOI:1007/978-981-99-4071-4_18.
CrossRef - Suryawanshi V, Adivarekar S, Bajaj K, Badami R. Comparative study of regularization techniques for VGG16, VGG19 and ResNet-50 for plant disease detection. Paper Presented at:Internatinal Conference on Communication and Computational Technologies (ICCCT); January 28-29, 2023; Jaipur. https://doi.org/10.1007/978-981-99-3485-0_61.
CrossRef - Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Paper presented at: 25th International Conference on Neural Information Processing Systems (NIPS); December 3, 2012; New York. https://doi.org/10.1145/3065386.
CrossRef - He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Paper presented at: IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 27-30, 2016; Las Vegas. https://doi.org/10.1007/978-981-99-3485-0_61.
CrossRef - Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21:6. https://doi.org/10.1186/s12864-019-6413-7.
CrossRef - Hughes DP, Salathé M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. Sci Data. 2015;2:150051. https://doi.org/10.48550/arXiv.1511.08060.
- PlantVillage Dataset. Available at: https://data.mendeley.com/datasets/tywbtsjrjv/1. Accessed August 22, 2024.

