Introduction
Polygonatum verticillatum L. is a highly valued medicinal herb of Indian Himalayan region (IHR) and occurs from 2000 -3000 m asl. It is a member of family Asparagaceae. It is known as “Meda” in Sanskrit and “Salam Mishri” in Hindi. This herb yield important metabolites and is one of the constituents of many ayurvedic formulations1-3. Morphologically, plants are slender and generally, unbranched with leaves arranged in verticillaster manner. Leaves are lanceolate, and stem bear flowers near the base of leaves. Flower colour is generally pale yellow and greenish. The fruits are round berries that are initially green in colour and become red when ripe. It is a plant of high importance. However, unscientific exploitation by local traders and anthropogenic activities has resulted in habitat destruction of its natural populations, which has resulted in the loss of the different wild genetic stocks of this valuable plant species. It has been reported from Uttarakhand that plant is vulnerable due to overuse by pahemaceutical companies, less awareness about the plant among local people, habitat destruction and habitat fragmentation in addition to unscientific harvesting and other anthropogenic activities4,5. Hence, it is also listed in threatened plants category6,7. In this situation, supreme importance should be given for characterizing the existing germplasm at molecular level for its genetic diversity and conserving its diverse germplasm. Genetic diversity data can be useful in identifying the diverse accessions which can be selected for conservation and for breeding experiments on priority basis. Diversity data can also be used for better management of the germplasm. Further, molecular characterization should be carried out at genetic level to identify elite germplasm stocks. This will help maintain diverse germplasms in the future and consequently will help in their conservation and management. Molecular markers, specifically DNA markers, are the highly preferred for the genetic characterization of any type of germplasm and plant collection. There are various types of DNA markers such as RFLP, RAPD, AFLP, ISSR, SSR, and SNP. Of these, SSR markers are currently the most commonly used DNA markers due to their desirable features such as the potential to resolve heterozygosity, easy laboratory procedures, cross-transferable nature and evenness in genomes8-15. Hence, these markers are widely used in plant genetic diversity research.
Moreover, the cross-transferability of SSR markers across species and genera is of great importance for exploring plant species for which SSR markers are not available. Many researchers have utilized the SSR markers from closely related and distantly related plant species to characterize and study the genetic diversity of germplasms of other species7,12-15. Few biochemical and genetic studies have been conducted in different species of Polygonatum, and more such studies are required for proper utilization and maintenance of this important genetic resource. Researchers categorized Polygonatum as an important genus with immense potential16,17. This plant contains many vital compounds, which exhibits curative and health promoting effects. Some important chemical constituents have been isolated and identified in this species. The researchers have developed method for identifying adulterants in mixtures of P.verticillatum18.
As it is a threatened plant, some conservationists have tried to establish tissue culture methods for its rapid propagation and availability19. However, genetic characterization studies on this species are lacking. In other species of the genus Polygonatum, such investigations have been performed. In P. sibiricum, attempt has been made to explore genes involved in biosynthetic pathways20. Other researchers identified some phytochemicals and antioxidants from this species21. P. cyrtonema Hua, which is an endemic species of this genus in China, was evaluated for its genetic diversity and structure in Anhui with the help of SSRs and morphological traits22. Genetic diversity of P. multiflorum and P. odoratum was also explored by Chinese researchers23. Cross-species SSR markers were evaluated for their cross- transferability in P. verticillatum24. A comparative study on clones and genetic structure was also undertaken in Polygonatum25. Different DNA markers were used for studying the genetic similarity of three species of Polygonatum, including P. verticillatum, occurring in Poland26. In India only one study using ISSR markers was conducted in P. verticillatum 27. Therefore, In the present study, SSR markers of an alpine species, namely, Betula utilis (Betula) were checked for cross-transferability in P. verticillatum and unambiguously amplified markers were utilized to characterize and study the genetic diversity of P. verticillatum. The rationale for using Betula utilis SSR was overlapping habitat of both the species. Both these species occurs in temperate and alpine regions of Himalaya and it was assumed that at genetic level these may contain similar genomic regions which help in adaptation of these species in their habitats. The results of current research work can be important for future research pertaining to conservation, breeding and management of this species.
Materials and Methods
Plant Sampling and DNA Extraction
Our sampling sites included state of Himachal Pradesh and Union Territory of Kashmir, India. Leaf samples from twenty two accessions of P. verticillatum were collected from different regions of four districts of Himachal Pradesh and one district of Kashmir. Young and fresh leaf samples were collected in plastic bags containing silica gel and transported to the laboratory at room temperature. DNA isolation was performed according to the CTAB method28 using liquid nitrogen. A detailed description of accessions is given in Table 1.
Table 1: List of accessions characterized in the present study using cross-transferred SSR markers
|
S. No. |
Sample code |
Location |
District |
State |
|
1. |
JH |
Jhungi |
Mandi |
Himachal Pradesh |
|
2. |
JL |
Jalori |
Kullu |
Himachal Pradesh |
|
3. |
TU-1 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
4. |
TU-2 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
5. |
KP |
Kataula |
Mandi |
Himachal Pradesh |
|
6. |
KS |
Fatehpura |
Anantnag |
Kashmir |
|
7. |
TU-3 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
8. |
TU-4 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
9. |
SR-1 |
Sarahan |
Shimla |
Himachal Pradesh |
|
10. |
SR-2 |
Sarahan |
Shimla |
Himachal Pradesh |
|
11. |
SN |
Solang Nullah |
Kullu |
Himachal Pradesh |
|
12. |
JL-1 |
Jalori |
Kullu |
Himachal Pradesh |
|
13. |
KT-1 |
Kala Top |
Chamba |
Himachal Pradesh |
|
14. |
KT-2 |
Kala Top |
Chamba |
Himachal Pradesh |
|
15. |
TU-5 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
16. |
TU-6 |
Tunga Dhar |
Mandi |
Himachal Pradesh |
|
17. |
JL-1 |
Jalori |
Kullu |
Himachal Pradesh |
|
18. |
KTH-1 |
Kainthley |
Chamba |
Himachal Pradesh |
|
19. |
KTH-2 |
Kainthley |
Chamba |
Himachal Pradesh |
|
20. |
KTH-3 |
Kainthley |
Chamba |
Himachal Pradesh |
|
21. |
KTH-4 |
Kainthley |
Chamba |
Himachal Pradesh |
|
22. |
KTH-5 |
Kainthley |
Chamba |
Himachal Pradesh |
PCR Reactions
In total, twenty-five SSR primers were checked for amplification in a pooled DNA sample. Among the 25 SSR primers of B. utilis29, 13 SSR primers were clearly amplified. Finally, these 13 unambiguously amplified primers were chosen for the SSR diversity study. The final volume of SSR reaction mixture was consisted of 10 µl. The composition of this included 4.8 µl deionized water, 2.0 µl template DNA of 13 ng/ µl quantity, 0.5 µl of each forward and reverse primer with 5 µM concentration, 0.5 µl MgCl2 (25 mM), 1.0 µl 10 X buffer containing 10 mMTris-Hcl, 50 mMKcl having pH 8.3, 0.5 µl dNTP mix consisting of 0.2 mM each dATP, dGTP, dCTP and dTTP, and lastly 0.2 µl Taq polymerase with 5U/ µl. The PCR conditions were set as: First stage- 1 cycle of 5 min at 94 ºC, Scond stage- 35 cycles of 1 min at 94 ºC, 1 min at annealing temperature of each primer, 1 min at 72 ºC and third stage- 7 min at 72ºC. PCR amplified products were run on 3% agarose gel in 1 X TBE buffer for visualization of fragments, and size of each fragment was determined with 100 bp DNA ladder (Genei, Bangalore). Ethidium bromide dye was used for detecting DNA fragments. Photo of gel was taken with the help of gel documentation system (Bio-print, Vilber Eppendorf, France).
Data Analysis
For analysis, DNA bands detected in the agarose gel were manually scored. The clearly amplified alleles were considered for scoring. After scoring, a binary data file was created in an Excel sheet, and all downstream analyses were performed using this binary file. Polymorphism Information Content (PIC) was determined with the formula as per Botstein and his group30. PIC is the indicatore of the level of polymorphism which the samples exhibit and can be employed to select polymorphic samples and primers. Similarly, Marker Index (MI) is the indicator of the efficiency of a marker and can be used to differentiate most informative versus least informative markers. Cluster analysis was done using distance based method and Jaccards similarity coefficient with UPGMA was used to generate dendrogram with DARwin31. The Groupings shown in the dendrogram were observed, and inferences and interpretations were made to reach conclusions.
Results
SSR Data and Diversity Indices
Thirteen SSR primers generated clear prominant alleles. In total, 12 primers were polymorphic, and generated 42 alleles. The average value of allele was 3.5. The size of alleles ranged from 100 bp to 600 bp. A representative gel image is given in Figure 1.
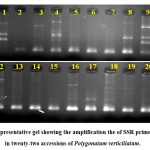 |
Figure 1: Representative gel showing the amplification the of SSR primer BUMS-5 in twenty-two accessions of Polygonatum verticillatum. |
The lowest number (2) of alleles were amplified by two primer pairs, i.e. BUMS-24 and BUMS-25 while the maximum number of alleles was 5 and amplified by two primers i.e. BUMS-03 and BUMS-15, as shown in Table 2. Maximum value of PIC was 0.500, exhibited by the primer BUMS-6 and the primer BUMS-22. Minimum value of PIC (0.375) was observed in case of primer BUMS-24. The mean PIC was 0.459. Maximum marker index was detected by the primer BUMS-15 (2.43), and lowest marker index was observed in case of the primer BUMS-24 (0.75). The mean marker index was 1.61. The dendrogram clustered studied accessions into three groups (Figure 2). Group-I consisted of two accessions i.e. SR-2 and KP. Group-II consisted of JA, JH, KS, SN, SR-1, TU-1, TU-2, TU-3, TU-4, TU-5 and TU-6. Group-III contained accessions KTH-1, KTH-2, KTH-3, KTH-4, KTH-5, JL, JL-1, KT-1 and KT-2.
Table 2: Features of primers used to characerize Polygonatum verticillatum accessions
|
S.No. |
Name of SSR primer |
Amlified or not (Yes/ No) |
No. of Alleles |
Sige range |
PIC |
MI |
|
1. |
BUMS-03 |
Yes |
5 |
160-500 |
0.404 |
2.02 |
|
2. |
BUMS-05 |
Yes |
4 |
150-700 |
0.490 |
1.96 |
|
3. |
BUMS-06 |
Yes |
2 |
200-330 |
0.5 |
1 |
|
4. |
BUMS-08 |
Yes |
4 |
170-500 |
0.456 |
1.82 |
|
5. |
BUMS-10 |
Yes |
4 |
180-420 |
0.493 |
1.97 |
|
6. |
BUMS-14 |
Yes |
1 |
400 |
– |
– |
|
7. |
BUMS-15 |
Yes |
5 |
150-500 |
0.486 |
2.43 |
|
8. |
BUMS-16 |
Yes |
3 |
150-500 |
0.444 |
1.33 |
|
9. |
BUMS-18 |
Yes |
3 |
200-500 |
0.454 |
1.36 |
|
10. |
BUMS-21 |
Yes |
4 |
100-400 |
0.479 |
1.91 |
|
11. |
BUMS-22 |
Yes |
4 |
190-600 |
0.5 |
2 |
|
12. |
BUMS-24 |
Yes |
2 |
160-250 |
0.375 |
0.75 |
|
13. |
BUMS-25 |
Yes |
2 |
220-370 |
0.433 |
0.867 |
|
Mean |
3.5* |
– |
0.459 |
1.61 |
||
PIC: Polymorphism Information Content, MI: Marker Index, *BUMS-14 was not included for finding the mean value of No. of Alleles as it was monomorphic.
Discussion
Genetic diversity indicates allele polymorphism in an observed plant species and can be an excellent measure for estimating the dynamics of alleles through time. Change in alleles of the existing populations may be driven by forces such as selection, habitat destruction, overexploitation, anthropogenic and geographical disturbances. On the other hand, the detected polymorphisms can be utilized to manage and manipulate the existing germplasm for improvement through different plant breeding approaches. Hence, detecting DNA diversity is a prerequisite for different types of research. Previously, Suyal and her co-authors investigated the morphological, phytochemical and genetic diversity of P. verticillatum from Uttarakhand using ISSR markers26. However, there study did not include any sample from Kashmir or Himachal Pradesh. In addition, ISSR markers are supposed to be dominant markers and may not reveal genetic aspects as the SSR markers can. Furthermore, they also suggested a high genetic diversity in the studied samples. In the present study, SSR markers of a distantly related plants species namely, B. utilis were successfully employed. Among 12 markers, 11 had PIC value greater than 4. Hence, it is suggested that the markers used in the present study can also be helpful for profiling and assessing population structure at larger levels with more samples. In the past, several authors have studied different species of Polygonatom, but the lines of work and methods used differed. Sheng and his group analyzed P. cyrtonema and showed normal levels of genetic diversity, and accessions clustered into three distinct genetic groups21. Other workers evaluated the phenotypic variation and population genetic structure of P. multiflorum and P. odoratum and their results revealed the effect of light availability to flowering intensity and population structure22. Sharma with his co-authors checked the cross- transferability of SSR markers of T. govanianum in P. verticillatum and oberseved that 10 SSRs showed reliable amplification23. Chung and his team used twenty-one allozyme loci in P. stenophyllum and P. inflatum24. Their data suggest that populations of P. stenophyllum have been mainly founded by a single seed or rhizome by river water or by few seeds, whereas populations of P. inflatum would have been established through multiple, repeated seedling recruitment. Szczecińska with his group determined the genetic similarity of three species i.e. P. multiflorum, P. odoratum and P. verticillatum, and concluded that P. verticillatum showed no considerable diversity25. When compared to some earlier studies which were done on different Himalayan herbs by various authors, it was found that mean value (0.459) of PIC and mean MI value (1.61) was higher than observed in Trillium govanianum by Dhyani and his co-workers32. On the other, Rana et al. observed 0.513 value of PIC which was higher than observed by us in current study33. A high level of genetic diversity was reported in another Himalayan herb Rheum australefrom western Himalayan region34. Pant et al. also recorded 0.46 value of PIC which was almost equal to the value detected by us herein35. These diversity values indicated that considerable genetic diversity exists in P. verticillatum which is comparable to diversity detected in other Himalayan herbs.
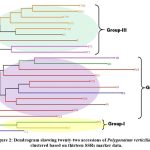 |
Figure 2: Dendrogram showing twenty-two accessions of Polygonatum verticillatum clustered based on thirteen SSRs marker data. |
The diversity detected in the present study was high, and the dendrogram clustered all the twenty-two accessions into three groups. The samples were grouped according to geographical locations; however, few samples were mixed within different geographical locations. Furthermore, groupings of the dendrogram revealed that the samples from Chamba district (KTH-1, KTH-2, KTH-3, KTH-4, KTH-5, KT-1 and KT-2) of Himachal Pradesh were conserved and grouped into a single group with two accessions (JL and JL-1) from Kullu district. Other accessions seemed to be mixed within the three groups; however, the accessions of disctrict Mandi (JH, KS, SN, SR-1, TU-1, TU-2, TU-3, TU-4, TU-5 and TU-6) remained in one group except for KP. These exceptions in clustering may be attributed to low number of markers used. However, it is interesting that the distantly related SSR markers have the ability to distinguish and generate enough polymorphisms for genetic diversity studies in P. verticillatum. Group-II also showed subclustering to some extent, and three sub-groups were detected. Except for one accession from Kashmir and one accession from Shimla, the other accessions from district Mandi and district Kullu almost grouped in single groups, which indicate that every population has some conserved alleles which resulted in such clustering. Based on this diversity data, the populations belonging to diverse accessions such as TU4, SR2, KP and KS can be selected for conservation in their natural strands. Moreover, the germplasm of identified populations can be raised through tissue culture techniques for its large scale propagation for even commercial cultivation. Furthermore, a detailed study with a large sample size could be helpful. The limited number of SSR markers and small sample size can be the limitation of the present study; however, the results are encouraging. In the future, studies with large number of samples and more SSR markers are needed for the clarity of allele arrangements in different populations of this species.
Prospective Conservation and Management Plans
At present, natural populations of P. verticillatum are found in patches of Indian Himalayan regions and extracted from these natural populations to meet the demand for the different purposes, such as medicine production and other dietary items by folklore and industrial systems. Hence, this threatened plant species need conservation and management for its proper utilization. Urgent attention and steps are required in this direction to safeguard this important germplasm resource. For in-situ conservation, cultivation in native regions with the help of neighboring dwellers and forest departments can be highly beneficial. This can help increase its population size and be utilized economically in a sustainable way. For ex-situ conservation, introduction and propagation at different locations with the help of scientific advisors from nearby organizations is necessary. Tissue culture strategies should be employed for large scale plantations. Furthermore, it can also be introduced in those Himalayan regions that are suitable for its natural growth so that its distribution may be widened. Natural extraction should be monitored scientifically to ensure the collection of roots and plant parts at appropriate stage of growth. At least extraction should not be done before the seed set. Government bodies and other organizations involved in the conservation of flora and environment needs to take care of these types of activities. Proper documentation of the quantity of the plant material harvested and supplied should be maintained at the native production site.
Conclusion
In conclusion, the present study reported the diversity of P. verticillatum using cross-transferred SSR markers of B.utilis. The results of this study showed that considerably high genetic diversity prevails in the populations of P. verticillatum in IHR. However, record of the population distribution and spreading trends in this region are not available. Proper conservation and management of this plant species is urgently needed for sustainable utilization. Large-scale studies involving maximum populations of IHR should be initiated for the identification of diverse accessions for improvement in the future. The SSRs used in this study can help exploring genetic variations and identifying important alleles in P. verticillatum.
Acknowledgement
N-PDF to Dr. Vikas Sharma from the SERB (DST) Government of India is highly acknowledged. Authors are thankful to Chancellor, Vice Chancellor and Registrar of Sant Baba Bhag Singh University for the creation of necessary laboratory facility in the University.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval” in this section
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Authors’ Contribution
PDS done sampling, experimental work and VS analyzed the data and wrote manuscript.
References
- Balkrishna A., Srivastava A., Mishra R.K., Patel S.P., Vashistha R.K., Singh A. Astavarga plants- threatened medicinal herbs of the North-West Himalaya. Int J Medicinal Aromatic Plants, 2012; (2) 661-76.
- Khan H., Saeed M., Khan M.A., Dar A., Khan I. The antinociceptive activity of Polygonatum verticillatum rhizomes in pain models. J Ethnopharmacology. 2010;127(2):521-7.
CrossRef - Virk J.K., Bansal P., Gupta V., Kumar S., Singh R. Lack of pharmacological basis of substitution of an endangered plant group Ashtawarga–A significant ingredient of polyherbal formulations. Am J Phytomed Clin Ther, 2015; 2:690-712.
- Bhatt D., Kumar R., Tewari L.M., Joshi G. C. Polygonatum cirrhifolium Royle and Polygonatum verticillatum (L.) Allioni: Status assessment and medicinal uses in Uttarakhand, India. Journal of Medicinal Plant Research, 2014; 8(5): 253-259.
CrossRef - Lohani N., Tewari1 L.M., Kumar R, Joshi G.C., Chandra J., Kishore K., Kumar S., Upreti B.M. Population studies, habitat assessment and threat categorization of Polygonatum verticillatum (L.) Allioni in Kumaun Himalaya. J Ecol Nat Environ, 2013; 5(5): 74-82.
CrossRef - Tiwari T., Chaturvedi P. Polygonatum verticillatum: A Highly Valued “Astavarga’’ Medicinal Herb of North West Himalaya. Med Plants, 2016; 8 (2): 97-103.
CrossRef - Suyal R., Jugran A.K., Rawal R.S., Bhatt I.D. Morphological, phytochemical and genetic diversity of threatened Polygonatum verticillatum (L.) All. populations of different altitudes and habitat types in Himalayan region. Physiol Mol Biol Plants, 2021; 27(8):1795-809.
CrossRef - Sharma R.K., Gupta P., Sharma V., Sood A., Mohapatra T., Ahuja P.S. Evaluation of rice and sugarcane SSR markers for phylogenetic and genetic diversity analyses in bamboo. Genome, 2008; 51:91–103.
CrossRef - Zhou Q., Cai H., Liu Z., Yuan L., Yang L., Yang T., Li B., Li P. Development of genomic resources for Wenchengia alternifolia (Lamiaceae) based on genome skimming data. Plant Diversity. 2022; 44:542-551.
CrossRef - Kaur K., Sharma V., Singh V., Wani M.S., Gupta R.C. Development of novel SSR markers for evaluation of genetic diversity and population structure in Tribulus terrestris L. (Zygophyllaceae). 3Biotech, 2016; 6:156.
CrossRef - Koul P.M., Sharma V., Rana M., Chahota R.K., Kumar S., Sharma T.R. Genetic Structure and Interrelationships in Lentil Species Using Morphological and SSR Markers. 3Biotech, 2017; 7:83.
CrossRef - Sharma H., Kumar P., Singh A., Aggarwal K., Roy J., Sharma V., Rawat S. Development of polymorphic EST-SSR markers and their applicability in genetic diversity evaluation in Rhododendron arboreum. Mol Biol Reports, 2020; 47(4):2447-2457 http://doi.org/10.1007/s11033-020-05300-1.
CrossRef - Sharma H., Kumar R., Sharma V., Kumar V., Bhardwaj P., Ahuja P.S., Sharma R.K. Identification and cross-species transferability of 112 novel unigene-derived microsatellite markers in tea (Camellia sinensis) Am J Bot, 2011; 98:e133-e138.
CrossRef - Aiello D., Ferradini N., Torelli L., Volpi C., Lambalk J., Russi L., Albertini E. Evaluation of Cross-Species Transferability of SSR Markers in Foeniculum vulgare. Plants, 2020;9:175.
CrossRef - Bhandawat A., Sharma V., Sharma H., Sood A., Sharma R.K. Development and cross transferability of functionally relevant microsatellite markers in D. latiflorus and related bamboo species. J Genetics, 2014; DOI 10.1007/s12041-014-0377-9.
CrossRef - Rana A., Kumar A., Savita. Polygonatum species and strategies for sustainable harvesting. The Indian Forester, 2017; 143(3).
- Zhao X., Li J. Chemical constituents of the genus Polygonatum and their role in medicinal treatment. Nat Prod Comm, 2015; 10(4):1934578X1501000439.
CrossRef - Virk J.K., Kumar S., Singh R., Tripathi A.C., Saraf S.K., Gupta V., Bansal P. Isolation and characterization of quinine from Polygonatum verticillatum: A new marker approach to identify substitution and adulteration. J Advanced Pharma Tech Res, 2016; 7(4):153.
CrossRef - Bisht S., Bisht N.S., Bhandari S. In vitro micropropagation in Polygonatum verticillatum (L.) All. an important threatened medicinal herb of Northern India. Physiol Mol Bio Plants, 2012; 18:89-93.
CrossRef - Wang S., Wang B., Hua W., Niu J., Dang K., Qiang Y., Wang Z. De novo assembly and analysis of Polygonatum sibiricum transcriptome and identification of genes involved in polysaccharide biosynthesis. Int J Mol Sciences, 2017; 18(9):1950.
CrossRef - Suyal R., Rawat S., Rawal R.S., Bhatt I.D. Variability in morphology, phytochemicals, and antioxidants in Polygonatum verticillatum (L.) All. populations under different altitudes and habitat conditions in Western Himalaya, India. Environ Monit and Assess, 2019; 191:1-8.
CrossRef - Sheng Y., Chen L., Li X., Shao J. Analysis of the genetic structure and morphology of Polygonatum cyrtonema in Anhui Province, eastern China revealed three distinct genetic groups. Nordic J Bot, 2020; 38(2).
CrossRef - Karpavičienė B., Mlečkaitė E.G. Response of Polygonatum multiflorum and P. odoratum morphological characteristics and population structure to variation in environmental factors. Botanica, 2019; 25(2):111-20.
CrossRef - Sharma V., Wani M.S., Singh V., Kaur K., Gupta R.C. Development and Characterization of Novel Microsatellite Markers in Trillium govanianum– a Threatened Plant Species from North-Western Himalaya. 3Biotech, 2017; 7:190.
CrossRef - Chung M.Y., López-Pujol J., Chung J.M., Kim K.J., Chung M.G. Contrasting levels of clonal and within-population genetic diversity between the 2 ecologically different herbs Polygonatum stenophyllum and Polygonatum inflatum (Liliaceae). J heredity, 2014; 105(5):690-701.
CrossRef - Szczecińska M., Sawicki J., Polok K., Hołdyński C., Zieliński R. Comparison of three polygonatum species from poland based on dna markers. Annales Bot Fennici, 2006; 43: 379–388.
- Suyal R., Bahukhandi A., Bhatt I.D., Rawal R.S. Comparative Analysis of Biochemical Attributes of Genus Polygonatum in Western Himalaya. Nat Acad Sc letters,2021; 457-460. doi:https://doi.org/10.1007/s40009-020-01028-5
CrossRef - Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus, 1990; 12(1):13.
CrossRef - Wani M.S., Gupta R.C., Munshi A.H., Sharma V. Development and Characterization of SSR markers in Himalayan tree species Betula utilis D. Don. J Forestry Res, 2018; 31: 1453-1460 doi.org/10.1007/s11676-019-00932-x.
CrossRef - Botstein D., White K.L., Skolnick M., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genet, 1980;32: 314–33l.
CrossRef - Perrier X., Jacquemoud-Collet, J.P. DARwin software http://darwin.cirad.fr/darwin, 2006.
- Dhyani P., Sharma B., Singh P., Masand M., Seth R., Sharma R.K. Genome-wide discovery of microsatellite markers and, population genetic diversity inferences revealed high anthropogenic pressure on endemic populations of Trillium govanianum. Scientific Reports, 2020; 15: 11269.
CrossRef - Singh H.C., Tiwari V., Tiwari A., Rana T.S. Development of EST-SSR markers in Bergenia ciliata using de novo transcriptome sequencing. Genome, 2024; 67(4), doi.org/10.1139/gen-2023-0059.
CrossRef - Kumar A., Bisht Y., Rautela K., Arun K. Jugran A.K., Bhatt I.D., Bargali S.S.Morphological and genetic diversity assessment of Rheum australe D. Don – A high value medicinal herb from the Himalaya, and implications for conservation strategies. South African Journal of Botany,2023; 163:620-629.
CrossRef - Oliya B.K., Maharjan L., Pant B. Genetic diversity and population structure analysis of Paris polyphylla Sm. revealed by SSR marker. Heliyon, 2023; DOI10.1016/i.heliyon.2023.e18230.
CrossRef

