Introduction
Barnyard millet (Echinochloa sp.) belongs to the sub-family panicoideae, family poaceae consisted of Japanese barnyard millet (E. esculenta (A. Braun) H. Scholz) and Indian barnyard millet (E. frumentacea Link). It is cultivated for food and feed purpose in Japan, Korea, the north-eastern parts of China and India, Pakistan and Nepal.1 It is having good nutritional properties as it is a good source of highly digestible proteins, and insoluble fractions of fibres.2 It is also effective for diabetic patients because of its ability to reduce the blood glucose levels.3 Though a lot of progress has been done in other major cereals and millets at molecular level, but in barnyard millet very few nucleotide sequences (41) were available in the NCBI database even in the present next generation sequencing era. There are no genomic microsatellites available for barnyard millet.
The discovery of conserved synteny in minor cereals with reference to the major cereals like rice, maize and wheat is very important to identify useful alleles of important agro-morphological traits. Availability of molecular markers such as microsatellite markers or simple sequence repeat markers (SSRs) and single nucleotide polymorphic markers (SNPs) is very essential for barnyard millet crop improvement programmes. Though the SSRs are highly polymorphic and preferred, but they have limitations such as they are often tedious, costly cloning and enrichment procedures required for their development.4,5,6 Microsatellites or simple sequence repeat (SSR) markers have been useful for molecular breeders and geneticists to link phenotypic and genotypic variation and also popular because of their abundance and amenability to high throughput screening. The genomic microsatellites offer few advantages over EST based SSRs like higher percentage of polymorphism.7 In this concern, microsatellites developed from expressed sequence tags (ESTs), popularly known as EST–SSRs or genic microsatellites have become attractive forces towards identification of EST based SSRs and SNPs for different omics applications of different crops. The In-Silico mining of EST-SSRs was reported in several crops like finger millet8, citrus9, coffee10 and other cereals,11 where as in-silico mining of SNPs was reported in crops like sorghum12 13 Babu et al8 did in-silico analysis of finger millet EST sequences available at NCBI and developed a total of 545 primer pairs from the ESTs of finger millet. Girma et al.,13 reported a total of 12,421 putative SNPs in sorghum from 2,921 contiguous transcripts leading to an average SNP interval of one putative SNP for every 275.26 bp.
However, in barnyard millet till now there no reports on identification of EST based SSRs and SNPs. With this aim the present study was conducted with the objective of 1) In-silico mining, type and frequency analysis of EST based SSRs, 2) In-silico mining of EST based SNPs, and 3) comparative genomic analysis of EST sequences to identify the putative orthologous regions in barnyard millet.
Materials and Methods
EST data mining
A total of 41 EST sequences publicly available at NCBI (GenBank) database for barnyard millet were downloaded in FASTA format for SSR and SNP analysis. The present study was conducted at ICAR-Vivekananda Parvateeya Krishi Anusanthan Sansthan, Almora, Uttarakhand in the year 2014-15. The identification of SSR in the EST sequences was done using a “websat” software 14 tool which uses the primer3 software as a basic programme. The riteria given for searching the simple sequence repeats was as follows. The criteria for mono-, di-, tri-, tetra-, penta-, and hexa- repeats did with minimum of 6, 5, 4, 3, 3 and 3 respectively. The main parameters for primer designing were: GC content of 50–60 %, annealing temperature (Tm) of 50–600C.
Strategy of comparative analysis of EST sequences of barnyard millet
The EST sequences of barnyard millet were retrieved from the NCBI website. To identify the homologous and orthologous genomic regions between barnyard millet with other crop genomes, these EST sequences were used for BLASTn analysis at an E value more than 4e-13.
Mining of SNPs
The presence of SNPs was done using the online software tool HaploSNPer.15 The strategy followed for in-silico identification of SNPs was as followed. The barnyard millet EST sequences were retrieved from the NCBI database. Vector sequences were trim to remove contaminated EST sequences. Redundancy in retrieved ESTs was established through clustering using codoncode aligner (http://www.codoncode.com/aligner/). All ESTs of barnyard millet were included in one batch and one of the sequences was included in other batch as input. Like wise to find out the SNPs presence between barnyard millet with other crop genomes like rice, maize, barley was done. To minimize the inclusion of ESTs from paralogous genes, stringent clustering criteria were followed: at least 50 bases overlap and 95% identity between one end of a sequence to the other end. The presence of variations was identified in assembled contigs. A quality screen was performed in order to differentiate genuine SNPs from false ones using the method adopted from Picoult-Newberg et al.,16 and Hawken et al17
Results and Discussion
Mining, frequency and distribution of EST-SSRs in barnyard millet
Till August, 2014, a total of 41 EST sequences of barnyard millet were available in the NCBI website and were downloaded in FASTA format. The in-silico mining of EST based SSRs was performed using the online software tool websat 14 and identified the forward and reverse primers using the primer 3 software embedded in the websat software. The SSR mining was done using the parameters set for repeat motifs as: di-, tri- tetra- penta- and hexa- nucleotide repeat units with > 6, >4, >3, >3, and >3 respectively. As per the above parameters, a total of seven ESTs were found to contain the microsatellite repeats. The remaining 34 sequences were very short length and did not have microsatellite repeats. A total of 22 primer pairs were able to design from the seven EST sequences. Recently Gimode et al.,18 identified 10,327 SSR primers by next generation sequencing in finger millet. Among these EST sequences of our study, five consisted of two primer pairs, while the remaining two sequences (AB668984.1 and AB668983.1) had six primer pairs. The details of the SSR repeat motifs, primer pair sequences, length and expected product size were given in table 1. The number of primers identified is similar to the earlier reports in finger millet.8 Babu et al.,8 identified a total of 545 primer pairs and of which 32 EST sequences had more than two microsatellites.
Table 1: The frequency, type and distribution details of EST-SSR of barnyard millet.
|
GenBank No |
Primer | Repeat motif | Length | Tm (0C) | Start/ End |
Expected product size (bp) |
| AB668988.1 | 5’TACAGCTTCTGATGAGCAATGG3′ | (CAAA)3 | 22 | 60 | 2632 | 302 |
| 5’CATGTCAAGAGCGATATGATGC3′ | 22 | 60 | 2912 | |||
| 5’TATTCTTCGCCCTTGTGCTTTA3′ | (AGACGA)3 | 22 | 61 | 3547 | 304 | |
| 5’TGTTCTTATCCAGACGGGTAGC3′ | 22 | 60 | 3829 | |||
| AB668987.1 | 5’GTCACTTGCATCATCATCTGTT3′ | (TGTTT)3 | 22 | 57 | 579 | 371 |
| 5’CTTGTAGCAGTGGAAGAACCTC3′ | 22 | 58 | 928 | |||
| 5’CGACTCACTCACTTCTGACACC3′ | (TGCA)3 | 22 | 59 | 803 | 308 | |
| 5’AGTCTTTCCCCAAACCTGAAAT3′ | 22 | 60 | 1089 | |||
| AB668986.1 | 5’TACAGCTTCTGATGAGCAATGG3′ | (CAAA)3 | 22 | 60 | 2632 | 302 |
| 5’CATGTCAAGAGCGATATGATGC3′ | 22 | 60 | 2912 | |||
| 5’TATTCTTCGCCCTTGTGCTTTA3′ | (AGACGA)3 | 22 | 61 | 3547 | 304 | |
| 5’TGTTCTTATCCAGACGGGTAGC3′ | 22 | 60 | 3829 | |||
| AB668985.1 | 5’GTCACTTGCATCATCATCTGTT | (TGTTT)3 | 22 | 57 | 579 | 371 |
| 5’CTTGTAGCAGTGGAAGAACCTC3′ | 22 | 58 | 928 | |||
| 5’CGACTCACTCACTTCTGACACC3′ | (TGCA)3 | 22 | 59 | 803 | 308 | |
| 5’AGTCTTTCCCCAAACCTGAAAT3′ | 22 | 60 | 1089 | |||
| AB668984.1 | 5’AGTACCAACAACAACAACTGGC3′ | (GCCT)3 | 22 | 59 | 997 | 378 |
| 5’CTTTCGGCTCCTTCCTTGT3′ | 19 | 59 | 1356 | |||
| 5’AGTACCAACAACAACAACTGGC3′ | (TCGA)3 | 22 | 59 | 997 | 378 | |
| 5’CTTTCGGCTCCTTCCTTGT3′ | 19 | 59 | 1356 | |||
| 5’GCCACCGCACTAGAAATACC3′ | (CAC)5 | 20 | 59 | 1607 | 388 | |
| 5’TCGACTAGCAGATCCATGAAAG3′ | 22 | 59 | 1973 | |||
| 5’AACACTCCACACCACACCAC3′ | (AGA)5 | 20 | 59 | 1663 | 367 | |
| 5’TAAGACAGGATGCGAAACACAC3′ | 22 | 59 | 2008 | |||
| 5’ACCAAGGAACAGCAAGAAGAAG3′ | (TCCC)3 | 22 | 59 | 1690 | 340 | |
| 5’TAAGACAGGATGCGAAACACAC3′ | 22 | 60 | 2008 | |||
| 5’TACAGCTTCTGATGAGCAATGG3′ | (CAAA)3 | 22 | 60 | 4790 | 302 | |
| 5’CATGTCAAGAGCGATATGATGC3′ | 22 | 60 | 5070 | |||
| AB668983.1 | 5’GATACAGGAGAGGGAGAGGGAT3′ | (AGGGAG)3 | 22 | 59 | 257 | 397 |
| 5’GCTGATATGGCAGAGGAGAGAG3′ | 22 | 60 | 632 | |||
| 5’CTTGTTTCTTTTGATGAAGCCC3′ | (GCGG)3 | 22 | 60 | 1295 | 396 | |
| 5’TATTTCTAGTGCGGTGGTGTGA3′ | 22 | 60 | 1669 | |||
| 5’CACACTTCACACACCACAACAC3′ | (AGA)9 | 22 | 60 | 1729 | 372 | |
| 5’CACAATCAAGAACACAGAGGGA3′ | 22 | 60 | 2079 | |||
| 5’TGGCTGTAAGTCCATCACTTCTT3′ | (TGTTT)5 | 22 | 60 | 2958 | 327 | |
| 5’GGTGTGGTGTCAGAACTCAAAA3′ | 22 | 60 | 3263 | |||
| 5’TGTTCATCGTGGTGTCTTCTTC3′ | (GT)10 | 22 | 60 | 3076 | 310 | |
| 5’GAACCTCACCCTCTCATACCTG3′ | 22 | 59 | 3364 | |||
| 5’TTTTGAGTTCTGACACCACACC3′ | (TGCA)3 | 22 | 60 | 3263 | 114 | |
| 5’CCTCTCATACCTGTCTCCCATC3′ | 22 | 59 | 3355 | |||
| AB668982.1 | 5’GTCACTTGCATCATCATCTGTT3′ | (TGTTT)3 | 22 | 57 | 1944 | 371 |
| 5’GAGGTTCTTCCACTGCTACAAG3′ | 22 | 58 | 2293 | |||
| 5’CGACTCACTCACTTCTGACACC3′ | (TGCA)3 | 22 | 59 | 2168 | 308 | |
| 5’AGTCTTTCCCCAAACCTGAAAT3′ | 22 | 60 | 2454 |
The percentage of different SSR repeat motifs identified in barnyard millet EST sequences has been given in table. The most frequent SSR repeats found to be tetra- nucleotide repeats representing of 50% of the total detected SSRs. The frequency and percentage of different microsatellite repeat motifs in the barnyard millet crop was given in table 2. However, either di- or trimeric nucleotide repeats were the most frequent repeat motifs in most crop species as reported in earlier findings.16 Gimode et al.,18 also found in finger millet that di-repeats were most frequent followed by tri and tetra repeat motifs. Babu et al.,8 found GA as the most common dimer repeat motif, while in case of trimeric SSRs, it was CGG in finger millet. Reddy et al.,16 also found GA was the most common motif, followed by AG repeat motif in finger millet. This variation may be due to that very less number of EST sequences were available in the public domain NCBI. In the present study, tetra nucleotide repeats were most frequent followed by the penta- and hexa- nucleotide repeats. Guanmei et al.,18 mined the EST based SSRs from Torreya grandis and found that di-repeat motifs are most frequent. They found that AAG/CCT tri repeat motif is the most prominent repeat motif.
Among the dimeric SSRs, GT was found to be the most common repeat motif. However, in case of finger millet GA/AG was the most common dinucleotide repeat8, 19 and in Poaceae crops AG/CT and GA/TC were the most frequent.20 However, GT repeat motif was found to be most frequent in algal species21 similar to the present study results. Among the tri repeat SSR motifs, AGG was the most common repeat motif, however, in case of finger millet and rice crops CGG was the common trinucleotide repeat motif. In rice CCG/CGG was the predominant tri- repeat motif.22 The most common tetra-, penta- and hexa nucleotide repeat motifs motifs were AGA, CAAA, TGTTT, AGACGA respectively. The graphical representation of the frequency of SSR motifs of ESTs of barnyard millet given in figure 1. The summary of the In-silico analysis of EST based SSRs in finger millet was depicted in table 3.
Table 2: The frequency and percentage of different microsatellite repeat motifs in the barnyard millet crop.
| SSR Repeat type |
Frequency |
Percentage |
| Di |
1 |
4.5 |
| Tri |
3 |
13.6 |
| Tetra |
11 |
50 |
| Penta |
4 |
18.2 |
| Hexa |
3 |
13.7 |
| Total |
22 |
100 |
Table 3: The summary of the In-silico analysis of EST based SSRs in finger millet.
| S. No | Parameters | Values |
| 1 | Total sequences | 41 |
| 2 | ESTs having microsatellite repeats | 7 |
| 3 | ESTs with no microsatellite repeats | 34 |
| 4 | ESTs with two primer pairs | 5 |
| 5 | ESTs with 6 primer pairs | 2 |
| 6 | Total number of SSRs | 22 |
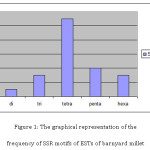 |
Figure 1: The graphical representation of the frequency of SSR motifs of ESTs of barnyard millet Click here to View figure |
In-Silico mining of single nucleotide polymorphisms (SNPs)
After retrieval of the barnyard EST sequences in FASTA format, the HaploSNPer software tool was used for identification of SNPs. The SNPs found in the EST sequences consisted of insertions/deletions (Indels), transitions and transversions. The present study is highly relevant in current scenario, where the need of effective molecular markers needed for various omics applications in crops like barnyard millet. Computational strategies for SNP discovery makes use of large number of sequences available in public database, in most cases an expressed sequence tags (EST) considered to be faster and cost effective than experimental procedures.15 In present study, the SNP mining was done in comparison of EST sequences of barnyard millet with rice, maize, sorghum.
Identification of SNPs among Barnyard millet sequences
All the EST sequences were grouped into 9 clusters. Out of the 9 clusters, the cluster 9 was found to have 1 potential SNP and 1 reliable SNP. This cluster has 2 haplotypes. The analysis of potential and reliable SNPs revealed that there are no indels (insertions and deletions). The potential SNPs have no transitions, but had one transversion (A\T). Among the reliable SNPs transversions was A\T and no transition was found. In cluster 9, the SNP was identified at 320 nucleotide place. The major allele was T, while the minor allele was A. Gimode et al.,18 found 23,285 non-homeologous SNPs in finger millet by next generation sequencing. The most abundant homeologous SNP was CT/AG (~62%) while CG was the rarest SNP at about 7.5%.
Identification of SNPs between Barnyard millet and rice genome
When barnyard millet EST sequences were used as input against the rice genome database no SNPs found with the default parameters as described in the materials and methods. However, the default parameter settings were changed as follows for identifying the SNPs. The E-value set as 1e-35, the similarity for this alignment was set to 90%. After changing the above parameters, it resulted in identification of 16 potential SNPs and 5 reliable SNPs. It was found that seven Indels were present in potential SNPs and no Indel was present in reliable SNPs. The potential SNPs found to contain one transition (A\G) and six transversions (A\T). Among the reliable SNPs one transition (A\G) and 3 transversions (A\C) were found. Nisha et al 23 developed single copy gene based 50K SNP chip in rice. This types of chips are very important not only in rice and also in similar related crops for their application in genomics platforms.
Identification of SNPs between Barnyard millet and maize genome
When barnyard millet EST sequences were used as input against the maize genome database no SNPs found with the default parameters as described in the materials and methods. However, the default parameter settings were changed as the similarity for this alignment is set to 90%, six SNPs were found. A total of six potential SNPs and one reliable SNP were found between barnyard millet and maize genome sequences. There were 3 Indels in potential SNPs and no Indel was found in reliable SNPs .The potential SNPs found to contain 3 transitions (C\T, and A\G), and no transversions. Among the reliable SNPs only one transition (A\G) was found. In cluster 1, the identified SNP was at 548 nucleotide place. The major allele A was found in 7 sequences, while the minor allele A was found in 2 sequences. One haplotype was found to contain the major allele, no haplotype was found to contain the minor allele. In cluster 3, the identified SNP was at 175 nucleotide place. Similarly Girma et al24 mines gene based SNPs in Sorghum bicolor as an cost effective genotyping assay. They identified 12,421 putative SNPs from 2,921 contiguous transcripts leading to an average SNP interval of one putative SNP for every 275.26 bp.
Comparative analysis of Barnyard millet EST sequences
Comparative genomics has facilitated the understanding of orthologues, and many fast evolving ‘orphan’ genes of unknown function and evolutionary history. In barnyard millets, comparative analysis provides an opportunity to study rapid genome changes, function of the newly isolated gene sequences and their further utilization in molecular breeding assisted crop improvement programmes for developing high yielding, stress resistant genotypes. In the present study, we compared the barnyard millet EST sequences with the rice genome database. It was found that most of the barnyard millet sequences showed homology to the rice chromosomal regions of 2, 5, 6, 8, 9 and 12. Similar type of study also conducted in finger millet, where they found high synteny between rice and finger millet25, 26 When we compared the similarity of barnyard millet ESTs with maize genome showed that more homology observed for Zea mays Waxy gene. The BLASTn analysis of barnyard millet EST sequences against the whole database in NCBI data base found to be more similar to GBSSI-S gene of Pannicum repens, Setaria italica, Pannicum milliacum Waxy gene, Colletotrichum eremochloae putative super oxide dismutase (SOD) gene. It needs many more EST sequences to find out the orthologous and homologous genes between barnyard millet and other cereals and millet species.
Conclusion
The research on genomics in barnyard millet lagging behind other millets and cereals, where there is a need of more focus towards identification of microsatellite markers. In the present work, a total of 22 primer pairs were developed from the ESTs. The most frequent SSR repeats found to be tetra- nucleotide repeats (50 percent) followed by the penta- and hexa- nucleotide repeats. The SNP mining of barnyard millet ESTs found to have 1 potential SNP and 1 reliable SNP and two haplotypes. Thus the identified twenty two microsatellite markers and SNPs can be effectively used for barnyard millet genomics applications to study diversity, and mapping aspects.
Acknowledgements
The authors thankful to the ICAR-VPKAS, Almora for providing financial assistance to carry out the work.
References
- Sood, S., Khulbe, R. K., Gupta, A., Agrawal, P. K., Upadhaya, H. D., and Bhatt, J. C Barnyard millet- A Potential Food and Feed Crop of Future. Plant Breeding. 2015;134:135-147.
CrossRef - Kantety, R, V., La Rota M., Matthews, D, E., and Sorrells, M, E Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol. 2002;48:501–510.
CrossRef - Maia, L. C., Souza, V. Q., Kopp, M, M., Carvalho, F. I. F., and Oliveira, A. C Tandem repeat distribution of gene transcripts in three plant families. Genet Mol Biol. 2009;32:1–12.
CrossRef - Poncet, V., Rondeau, M., Tranchant, C., Cayrel, A., Hamon, S., de Kochko, A., and Hamon, P SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Gen Genomics. 2006;276:436–449
CrossRef - Reddy, B. L. I. N., Lakshmi, N. M., and Sivaramakrishnan, S Identification and characterization of EST–SSRs in finger millet (Eleusine coracana (L.) Gaertn.). J Crop Sci Biotech. 2012;15(1):9–16.
CrossRef - Veena, S., Bharati, V. C., Rama, K. N., and Shantha kumar, G Physico-chemical and nutritional studies in barnyard millet. Karnataka J Agric Sci. 2005;18:101—105.
- Babu, B. K., Mathur, R. K., Kumar, P. N,, Ramajayam, D., Ravichandran, G., Venu , M. V. B Development, identification and validation of CAPS marker for SHELL trait which governs dura, pisifera and tenera fruit forms in oil palm (Elaeis guineensis Jacq.). PLoS ONE. 2017;12(2).
CrossRef - Zane, L., Bargelloni, L., and Patarnello, T Strategies for microsatellite isolation: a review. Mol Ecol. 2002;11:1–16.
CrossRef - Weising, K., Nybom, H., Wolff, K., and Kahl, G Application of DNA fingerprinting in plant sciences. DNA Fingerprinting in plants-principles, methods, and applications. CRC Press, Boca Raton. 2005;235–276.
- Squirrell, J., Hollingsworth, P. M., Woodhead, M., Russell, J., Lowe, A. J., Gibby, M., Powell, W. How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol. 2003;12:1339–1348.
CrossRef - Ugare, R., Chimmad, B., Naik, R., Bharati, P., and Itagi, S Glycemic index and significance of barnyard millet (Echinochloa frumentacae) in type II diabetics. J. Food Sci Technol. 2014;51:392—395.
CrossRef - Martins, W. S., Lucas, D. C. S., Neves, K. F. S., and Bertioli, D. J Web-Sat—A Web Software for microsatellite marker development. Bioinformation. 2009;3(6):282–283.
CrossRef - Victoria, F. C., da Maia Luciano, C., and de Oliveira Antonio, C. In silico comparative analysis of SSR markers in plants. BMC Plant Biol. 2011;11:15–30.
CrossRef - Girma, Y., Dadakhalandar, D., Fakrudin, B., Rajkumar., Sadik, A. W, A., Sheweta G., Suvarna, P., and Gurusiddesh, H. Mining of Gene-Based SNPs from Publicly Available ESTs and Their Conversion to Cost-Effective Genotyping Assay in Sorghum [Sorghum bicolor (L.) Moench]. J. Crop Sci Biotech. 2014;17(3):155–160.
CrossRef - Tang, J., Vosman, B., Voorrips, R. E., van der Linden, C. G., Leunissen, J. A. M QualitySNP: a pipeline for detecting single nucleotide polymorphisms and insertions/deletions in EST data from diploid and polyploid species BMC Bioinformatics. 2006;7:438 doi: 10.1186/1471-2105-7-438.
- Chen, C., Zhou, P., Choi, Y. A., Huang, S., and Gmitter, F. G. Jr Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet. 2006;112:1248–1257.
CrossRef - Babu, B. K., Dinesh Pandey., Agrawal, P. K., Salej Sood., and Anil Kumar In-silico mining, type and frequency analysis of genic microsatellites of finger millet (Eleusine coracana (L.) Gaertn.): a comparative genomic analysis of NBS–LRR regions of finger millet with rice. Mol Biol Rep. 2014;41(5):3081-3090.
CrossRef - Gimode, D., Odeny, D. A., de Villiers, E. P., Wanyonyi, S., Dida, M. M., Mneney, E. E. Identification of SNP and SSR Markers in Finger Millet Using Next Generation Sequencing Technologies. PLoS ONE. 2016;11(7).
CrossRef - Yu, J. K., Dake, T. M., Singh, S., Benscher, D., Li, W., Gill, B., and Sorrells, M. E Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome. 2004;47:805–818.
CrossRef - Hawken, R. J, Wesley, C. B, Sean, M., and Brian, P. D. An interactive bovine in silico SNP database (IBISS). Mammalian genome. 2004;15:819-827.
CrossRef - Deepika, S. S., and Asheesh, S. Mining of simple sequence repeats in chloroplast genome sequence of Cocos nucifera. Appl Res. 2015;1(2):51-54.
- Divya, S., Priyanka, G., Pragati, S., Neha, K., Siddharth, A., and Himanshu, S. In-silico single nucleotide polymorphisms (SNP) mining of Sorghum bicolor genome. Afr J Biotech. 2011;10(4):580-583.
- Nisha, S., Jayaswal, P. K., Kabita, P., Mandal, P., Vinod Kumar., Singh, B., Shefali Mishra, Yashi Singh, Renu Singh, Vandna Rai, Anita Gupta, Tilak Raj Sharma, Singh, N. K. Sci Rep. 2015;5:11600.
- Girma, Y., Doddamani, D., Fakrudin, B. et al. J. Crop Sci. Biotechnol. 2014;17: 155.
CrossRef - Babu, B. K., Dinesh Pandey, P. K. Agrawal,S. Sood, C. Chandrashekara, J. C. Bhatt, and Anil Kumar.. Comparative genomics and association mapping approaches for blast resistant genes in finger millet using SSRs. PLoS ONE. 2014;9(6).
CrossRef - Dida, M. M., R. S. Srinivasachary, J. L. Bennetzen, M. D. Gale, and K. M. Devos.. The genetic map of finger millet, Eleusine coracana. Theor. Appl. Genet. 2007;114:321–332.
CrossRef

