Introduction
The cactus pear (Opuntia spp.) is a plant genus native to arid and semi-arid regions of Mexico.1 It belongs to the family Cactaceae, which is consisting of about 1500 species30 and it is widely distributed throughout temperate, subtropical and cold regions (Feugang et al. 2006).In Morocco, the cactus pear is one of the most important crop plants in the semi-arid zones. It covers an area of about 120000 ha5 and it is represented mainly by the species Opuntia ficus indica. The accessions of this species are classified into three distinct populations: (i) the Christians’ nopal with prickly cladodes used as a field fence; (ii) the Muslims’ nopal with inermis cladodes used as a green fodder for cattle and (iii) the Moses’ nopal with large inermis cladodes that produce big pears.20 The cactus pear population is present throughout the country, from Eastern to Southern Morocco,18 with predominance in Guelmim-Sidi Ifni and Haouz-El Kelâa des Sraghnas regions.5 Increasing the cactus pear area of cultivation is one of the strategies of the Moroccan Ministry of Agriculture. The cactus pear is a tree with high economic and ecologic value and various uses. It is cultivated for human consumption since it is characterized by important nutritional components.42 The cactus pear has also many beneficial effects on health, including anti-cancer effect, anti-viral effect and anti-diabetic-effect among others.23 On the other hand, the cactus pear may be utilized as forage due to its nutritional characteristics; along this line, Andrade-Montemayor et al. (2011)4 reported that cactus cladodes could be used for goats feeding in semi-arid regions. The cactus pear was also successfully used for feeding Santa Inês lambs 13 and dromedary camel.15 Furthermore, it was reported that some cactus constituents could be extracted and used as food additives or for pharmaceutical and cosmetic purposes.32 The process of agricultural intensification and modern agricultural practices has led to genetic diversity degradation and gene pools loss.2-28 Determining the level of genetic diversity among and within plant populations is primordial for genetic resources conservation and breeding programs.24-28 The use of morphological descriptors is of great help for genetic diversity assessment.14 In the recent years, molecular markers have been proven powerful to determine the genetic diversity among and within species 12 and could be used as complementary methods to morphological descriptors.14 Along this line, inter-simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) markers have been widely used to assess the genetic diversity in many species. Both the techniques are characterized by their simplicity, rapidity, ability to show high polymorphism with no prior knowledge of the genome studied.36-41 In the recent years, several studies have been carried out on the genetic diversity of the cactus pear. For example, Valadez-Moctezuma et al. (2014)39 studied the genetic diversity within and among Mexican Opuntia species using RAPD and ISSR markers. In Italy, Labra et al. (2003) 27 evaluated the genetic diversity in Opuntia species with cpSSR and AFLP markers. Zoghlami et al. (2007)44 have assessed the genetic diversity of Opuntia ficus indica L. in Tunisia using RAPD markers. More recently, Bendhifi et al. (2013)6 published another work on the genetic diversity of the same species in Tunisia using morphological descriptors and RAPD markers. Despite the high importance of the cactus pear in Morocco, its genetic diversity has been scarcely studied. Therefore, the aim of this investigation was to assess the genetic diversity within and among Opuntia spp. from different regions of Morocco, using morphological descriptors as well as ISSR and RAPD markers.
Materials and Methods
Plant Material
In the following investigation, we used 124 Opuntia spp. accessions belonging to different species: O. ficus indica; O. megacantha; O. robusta; O. aequatorialis; O. dillenii; O. leucotricha and O. inermis (identified for the first time in Morocco; Table 1). The accessions are planted in the experimental site called Ain Nzagh of National Institute of Agronomic Research-Regional Research Centre of Settat, (INRA-CRRA Settat, Morocco). Originally, the accessions were collected from different geographical sites of Morocco as shown in Table 1 and Figure 1 then planted in the experimental site in June 2011. All the accessions have undergone the same culture edaphoclimatic conditions.
Morphological Analysis
In this study, 10 quantitative morphological traits including plant height, plant diameter, average number of cladodes per plant, cladode height, cladode width, index of cladode shape (calculated as the ration of one-year-old cladodes length divided by their width), cladode thickness, mean distance between areoles, average number of spines per areole and mean length of the longest spine per areole. Data were recorded from 4 randomly chosen plants (2 years old) for each accession. Morphological data were subjected to analysis of variance (ANOVA) and means were separated with the Student-Newman-Keuls test at a 0.05 probability level. A proximity matrix was generated using Squared Euclidean distance then clustering of accessions was performed using Ward’s method. All statistical analyses were performed with SPSS v 16.0 software (IBM, Chicago, IL, USA).
Table 1: Means (+ SD) of morphological traits in different accessions of Opuntia spp. in Morocco *Mean values with the same letters are not significantly different (p> 0.05) (-): Absence of spine.
| Species | Geographic origin | Accession | Plant height (cm) | Plant diameter (cm) | Average number of cladodes per plant | Cladode height (cm) | Cladode width (cm) | Index of cladode shape (cm) | Cladode thickness (cm) | Mean distance between areoles (cm) | Average number of spines per areole | Mean length of the longest spine per areole (cm) |
| O. ficus indica | Jamaât Riah | Ofi-B2 | 36.80 +3.16 abcd | 45.08 +1.92 abcdefghi | 3.00 +2.16 cdef | 26.63 +2.36 bcdefghijk | 13.63 +2.41bc | 0.51 + 0.09bcdef | 0.83 + 0.22abc | 2.70 + 0.36bcdefg | – | – |
| Jamaât Riah | Ofi-B3 | 37.10 +2.68 abcd | 47.13 +9.91 abcdefghi | 2.75 +0.96 cdef | 21.98 +5.54 defghijkl | 10.93 +2.44 bcd | 0.50 +0.06 bcdef | 0.68 +0.21 abc | 2.58 +0.15 cdefg | – | – | |
| Jamaât Riah | Ofi-C2 | 33.38 +8.84 abcd | 37.93 +16.12 abcdefghi | 3.00 +0.82 cdef | 21.80 +0.98 defghijkl | 13.10 +0.33 bc | 0.59 +0.01 bcdef | 0.98 +0.05 abc | 2.90 +0.08 bcdefg | – | – | |
| Jamaât Riah | Ofi-C3 | 36.25 +9.81 abcd | 37.00 +6.77 abcdefghi | 1.75 +0.50 ef | 24.88 +1.84 bcdefghijk | 13.25 +1.56 bc | 0.56 +0.02 bcdef | 0.73 +0.13 abc | 2.93 +0.50 bcdefg | – | – | |
| Jamaât Riah | Ofi-D1 | 29.50 +4.55 abcd | 28.38 +5.76 fghi | 2.50 +0.58 cdef | 20.08 +1.05 efghijkl | 11.35 +0.29 bcd | 0.57 +0.02 bcdef | 1.10 +0.00 abc | 2.45 +0.10 defg | – | – | |
| Jamaât Riah | Ofi-D2 | 30.13 +14.77 abcd | 32.38 +14.51 cdefghi | 2.00 +0.82def | 28.90 +3.53 bcdef | 13.19 +3.05 bc | 0.47 +0.05 bcdef | 1.20 +0.00 abc | 2.68 +0.49 bcdefg | – | – | |
| Jamaât Riah | Ofi-D3 | 36.38 +3.42 abcd | 51.38 +7.61 abcdefghi | 3.75 +0.50bcdef | 26.63 +3.38 bcdefghijk | 13.85 +1.64 bc | 0.52 +0.03 bcdef | 0.90 +0.47 abc | 2.68 +0.31 bcdefg | – | – | |
| Jamaât Riah | Ofi-E1 | 37.10 +10.54 abcd | 43.33 +4.19 abcdefghi | 2.25 +1.50def | 25.13 +2.78 bcdefghijk | 13.30 +1.72 bc | 0.62 +0.16bcde | 0.80 +0.29 abc | 3.05 +0.33 bcdefg | – | – | |
| Jamaât Riah | Ofi-E2 | 42.75 +4.86 abc | 52.25 +25.16 abcdefghi | 3.75 +1.89 bcdef | 27.43 +1.92 bcdefghij | 13.80 +0.68 bc | 0.50 +0.05 bcdef | 0.75 +0.41 abc | 2.90 +0.29 bcdefg | – | – | |
| Jamaât Riah | Ofi-F1 | 27.45 +12.12 abcd | 51.95 +32.28 abcdefghi | 3.50 +1.73 bcdef | 28.25 +3.77 bcdefg | 14.28 +1.81 bc | 0.49 +0.04 bcdef | 0.75 +0.41 abc | 3.75 +0.29 ab | – | – | |
| Jamaât Riah | Ofi-F2 | 37.25 +7.58 abcd | 66.00 +12.93 abc | 4.00 +1.15 bcdef | 28.15 +4.60bcdefgh | 14.25 +1.94 bc | 0.51 +0.04 bcdef | 0.78 +0.39 abc | 3.18 +0.62 bcdefg | – | – | |
| Jamaât Riah | Ofi-F3 | 31.88 +1.89 abcd | 34.00 +5.58 cdefghi | 2.25 +0.50def | 22.10 +2.68 defghijkl | 12.50 +0.71 bcd | 0.55 +0.05 bcdef | 0.60 +0.18 abc | 2.78 +0.26 bcdefg | – | – | |
| Jamaât Riah | Ofi-G1 | 41.25 +14.31 abcd | 37.28 +25.85 abcdefghi | 3.50 +2.08 bcdef | 27.55 +0.90 bcdefghij | 10.13 +2.25cd | 0.41 +0.08def | 1.05 +0.12 abc | 3.05 +0.29 bcdefg | – | – | |
| Jamaât Riah | Ofi-G2 | 32.63 +1.70 abcd | 30.13 +4.01 cdefghi | 1.50 +0.58 ef | 23.75 +3.77 cdefghijkl | 13.15 +2.21 bc | 0.55 +0.02 bcdef | 0.63 +0.13 abc | 2.80 +0.24 bcdefg | – | – | |
| Jamaât Riah | Ofi-H1 | 39.38 +6.63 abcd | 39.68 +1.89 abcdefghi | 2.50 +0.58 cdef | 26.45 +4.79 bcdefghijk | 13.73 +1.39 bc | 0.52 +0.08 bcdef | 0.98 +0.32 abc | 3.10 +0.20 bcdefg | – | – | |
| Jamaât Riah | Ofi-H2 | 30.00 +7.67 abcd | 35.75 +5.74 bcdefghi | 1.50 +0.58 ef | 24.00 +2.04 cdefghijk | 12.85 +0.53 bc | 0.53 +0.02 bcdef | 1.10 +0.16 abc | 2.65 +0.04 bcdefg | – | – | |
| Jamaât Riah | Ofi-H3 | 26.30 +6.53 abcd | 37.25 +2.33 abcdefghi | 2.00 +0.00def | 21.60 +1.96 defghijkl | 10.60 +1.55bcd | 0.48 +0.03 bcdef | 1.20 +0.08 abc | 2.60 +0.24 cdefg | – | – | |
| Jamaât Riah | Ofi-I2 | 29.88 +1.03 abcd | 29.00 +11.05 efghi | 1.75 +0.96 ef | 18.17 +6.18 ghijkl | 11.60 +1.10 bcd | 0.72 +0.25b | 0.70 +0.24 abc | 2.80 +0.43 bcdefg | – | – | |
| Jamaât Riah | Ofi-J1 | 30.93 +2.69 abcd | 33.00 +8.77 cdefghi | 2.50 +1.00 cdef | 21.33 +1.43 defghijkl | 10.43 +1.25cd | 0.49 +0.04 bcdef | 0.70 +0.22 abc | 2.73 +0.19 bcdefg | – | – | |
| Jamaât Riah | Ofi-J2 | 24.90 +10.25 bcd | 27.23 +11.98 fghi | 3.00 +0.82 cdef | 19.50 +0.41 fghijkl | 10.50 +1.22cd | 0.53 +0.05 bcdef | 0.90 +0.08 abc | 2.50 +0.41 defg | – | – | |
| Jamaât Riah | Ofi-J3 | 22.83 +4.28 cd | 30.50 +3.67 cdefghi | 2.75 +0.50cdef | 18.83 +1.09 fghijkl | 9.88 +2.29cd | 0.53 +0.14 bcdef | 0.98 +0.22 abc | 2.55 +0.13 cdefg | – | – | |
| Rhamna | Ofi-113 | 35.00 +3.34 abcd | 62.88 +15.93 abcdef | 4.25 +1.50 bcdef | 25.95 +3.09 bcdefghijk | 13.33 +1.48 bc | 0.51 +0.03 bcdef | 0.75 +0.41 abc | 2.75 +0.31 bcdefg | – | – | |
| Rhamna | Ofi-121 | 29.83 +2.17 abcd | 46.75 +10.20 abcdefghi | 6.25 +1.71ab | 25.28 +3.05 bcdefghijk | 12.58 +1.26 bcd | 0.50 +0.02 bcdef | 0.88 +0.43 abc | 2.90 +0.22 bcdefg | – | – | |
| Rhamna | Ofi-141 | 25.50 +0.00 bcd | 26.50 +3.27 ghi | 1.00 +0.00f | 16.30 +0.24 kl | 9.50 +0.41cd | 0.57 +0.05 bcdef | 0.90 +0.08 abc | 2.60 +0.16 cdefg | – | – | |
| Rhamna | Ofi-142 | 24.75 +6.50 bcd | 28.03 +4.01 fghi | 1.00 +0.00f | 22.60 +1.96 cdefghijkl | 12.85 +2.16 bc | 0.56 +0.05 bcdef | 1.15 +0.04 abc | 2.65 +0.53 bcdefg | – | – | |
| Rhamna | Ofi-151 | 33.93 +3.99 abcd | 45.45 +3.32 abcdefghi | 2.25 +1.26def | 26.70 +2.52 bcdefghijk | 12.00 +1.36 bcd | 0.45 +0.08 bcdef | 0.78 +0.22 abc | 3.00 +0.14 bcdefg | – | – | |
| Rhamna | Ofi-152 | 31.48 +5.12 abcd | 29.93 +1.34 defghi | 1.50 +0.58ef | 24.50 +3.39 bcdefghijk | 11.30 +2.43 bcd | 0.45 +0.04 bcdef | 0.80 +0.38 abc | 2.83 +0.46 bcdefg | – | – | |
| Rhamna | Ofi-153 | 27.10 +8.23 abcd | 32.78 +20.66 cdefghi | 2.00 +0.82def | 22.14 +3.60 defghijkl | 11.29 +2.47 bcd | 0.47 +0.05 bcdef | 1.00 +0.24 abc | 3.23 +0.66 bcdefg | – | – | |
| Rhamna | Ofi-161 | 26.28 +5.33 abcd | 31.33 +4.49 cdefghi | 1.25 +0.50f | 20.43 +3.81 efghijkl | 11.60 +2.05 bcd | 0.57 +0.06 bcdef | 0.78 +0.36 abc | 2.78 +0.32 bcdefg | – | – | |
| Rhamna | Ofi-162 | 27.25 +2.30 abcd | 43.05 +11.40 abcdefghi | 2.50 +0.58cdef | 22.37 +1.69 cdefghijkl | 12.77 +0.38 bc | 0.57 +0.03 bcdef | 0.80 +0.28 abc | 2.80 +0.14 bcdefg | – | – | |
| Rhamna | Ofi-163 | 42.13 +5.39 abcd | 71.38 +5.02 a | 4.75 +1.89bcde | 29.13 +4.21 bcdef | 14.53 +1.75 bc | 0.50 +0.05 bcdef | 0.88 +0.17 abc | 2.85 +0.87 bcdefg | – | – | |
| Rhamna | Ofi-1101 | 38.00 +4.32 abcd | 36.50 +7.08 bcdefghi | 2.25 +0.50def | 27.70 +2.95 bcdefghi | 13.48 +1.19 bc | 0.49 +0.04 bcdef | 0.95 +0.33 abc | 3.00 +0.56 bcdefg | – | – | |
| Ouardigha | Ofi-211 | 25.38 +9.62 bcd | 24.98 +9.72 ghi | 1.00 +0.00f | 22.25 +1.02 defghijkl | 12.00 +0.00 bcd | 0.54 +0.02 bcdef | 1.15 +0.12 abc | 2.40 +0.08 efg | – | – | |
| Ouardigha | Ofi-212 | 26.63 +3.68 abcd | 36.00 +6.16 bcdefghi | 1.50 +0.58 ef | 24.40 +2.57 bcdefghijk | 12.17 +1.03 bcd | 0.50 +0.05 bcdef | 1.13 +0.17 abc | 2.93 +0.45 bcdefg | – | – | |
| Ouardigha | Ofi-231 | 29.60 +3.93 abcd | 42.13 +12.68 abcdefghi | 2.50 +0.58 cdef | 21.10 +4.66 defghijkl | 11.05 +3.14 bcd | 0.52 +0.04 bcdef | 0.58 +0.21 abc | 2.53 +0.62 defg | – | – | |
| Ouardigha | Ofi-232 | 28.25 +3.97 abcd | 31.00 +4.32 cdefghi | 2.00 +0.82def | 18.00 +1.63 ghijkl | 10.30 +0.24cd | 0.57 +0.06 bcdef | 0.70 +0.16 abc | 2.80 +0.16 bcdefg | – | – | |
| Ouardigha | Ofi-233 | 24.13 +9.08 bcd | 29.00 +5.55 efghi | 1.75 +0.96 ef | 18.57 +1.41 fghijkl | 9.10 +0.65cd | 0.49 +0.02 bcdef | 0.73 +0.12 abc | 2.57 +0.49 cdefg | – | – | |
| Ouardigha | Ofi-261 | 22.30 +3.19 cd | 29.45 +6.44 defghi | 1.00 +0.00f | 19.60 +3.59 fghijkl | 10.65 +1.67 bcd | 0.54 +0.02 bcdef | 0.85 +0.12 abc | 2.70 +0.16 bcdefg | – | – | |
| Ouardigha | Ofi-262 | 26.70 +3.85 abcd | 35.63 +4.17 bcdefghi | 1.50 +0.58 ef | 22.81 +3.35 cdefghijkl | 11.43 +0.52 bcd | 0.49 +0.03 bcdef | 0.97 +0.34 abc | 2.73 +0.21 bcdefg | – | – | |
| Ouardigha | Ofi-263 | 39.13 +4.21 abcd | 44.60 +4.33 abcdefghi | 1.75 +0.50 ef | 30.45 +1.56abcde | 13.50 +0.71 bc | 0.50 +0.12 bcdef | 0.93 +0.46 abc | 3.15 +0.24 bcdefg | – | – | |
| Ouardigha | Ofi-264 | 28.33 + 4.44 abcd | 36.88 + 9.19 abcdefghi | 2.50 +1.00 cdef | 23.90 +1.34 cdefghijk | 11.27 +0.56 bcd | 0.47 +0.03 bcdef | 0.47+0.10 c | 2.40 + 0.26 efg | – | – | |
| Ouardigha | Ofi-265 | 24.25 +0.61 bcd | 35.25 +5.92 bcdefghi | 3.00 +0.82 cdef | 23.20 +0.16 cdefghijkl | 10.20 +0.16cd | 0.43 +0.08cdef | 1.10 +0.08 abc | 2.90 +0.73 bcdefg | – | – | |
| Ouardigha | Ofi-266 | 29.88 + 5.02 abcd | 27.75 +1.32 fghi | 1.50 +0.58 ef | 27.23 +6.65 bcdefghij | 13.77 +1.45 bc | 0.52 +0.06 bcdef | 0.77 +0.09 abc | 3.33 +0.24bcdef | – | – | |
| Ouardigha | Ofi-271 | 27.17 +7.71 abcd | 28.83 +8.66 efghi | 1.50 +0.58 ef | 26.20 +0.82 bcdefghijk | 12.45 +0.37 bcd | 0.47 +0.00 bcdef | 1.05 +0.04 abc | 3.00 +0.00 bcdefg | – | – | |
| Ouardigha | Ofi-272 | 35.88 +11.35 abcd | 30.75 +8.07 cdefghi | 2.00 +0.82def | 22.48 +3.26 cdefghijkl | 12.18 +0.96 bcd | 0.54 +0.06 bcdef | 1.03 +0.10 abc | 2.68 +0.28 bcdefg | – | – | |
| Ouardigha | Ofi-273 | 24.33 +5.09 bcd | 27.38 +12.15 fghi | 1.50 +0.58 ef | 17.53 +2.50 hijkl | 9.87 +0.26cd | 0.57 +0.10 bcdef | 0.63 +0.05 abc | 2.47 +0.39 defg | – | – | |
| Ouardigha | Ofi-274 | 28.38 +6.80 abcd | 35.00 +9.70 bcdefghi | 2.00 +0.82def | 19.73 +6.88 fghijkl | 10.25 +1.19cd | 0.56 +0.18 bcdef | 0.73 +0.10 abc | 2.43 +0.40 efg | – | – | |
| Ouardigha | Ofi-275 | 21.55 + 8.86 cd | 16.43 +3.51 i | 1.50 +0.58 ef | 18.55 +3.23 fghijkl | 12.30 +0.90 bcd | 0.67 +0.07bcd | 1.15 +0.04 abc | 2.70 +0.24 bcdefg | – | – | |
| Middle atlas | Ofi-311 | 35.68 +7.92 abcd | 50.88 +3.45 abcdefghi | 2.25 +0.50def | 24.15 +6.68 bcdefghijk | 11.65 +1.08 bcd | 0.50 +0.08 bcdef | 0.65 +0.10 abc | 2.53 +0.10 defg | – | – | |
| Middle atlas | Ofi-321 | 38.50 +19.07 abcd | 29.13 +12.80 efghi | 3.75 +0.96 bcdef | 23.47 +2.17 cdefghijkl | 12.87 +1.62 bc | 0.54 +0.03 bcdef | 0.70 +0.14 abc | 3.20 +0.14 bcdefg | – | – | |
| Middle atlas | Ofi-331 | 31.88 +8.23 abcd | 22.25 +1.66 ghi | 1.00 +0.00f | 13.70 +0.00 l | 10.70 +0.00 bcd | 0.65 +0.11 bcd | 0.80 +0.00 abc | 2.55 +0.04 cdefg | – | – | |
| Middle atlas | Ofi-341 | 30.63 +6.94 abcd | 31.38 +7.06 cdefghi | 1.50 +0.58 ef | 19.27 +6.73 fghijkl | 13.00 +0.50 bc | 0.51 +0.05 bcdef | 0.83 +0.17 abc | 3.13 +0.26 bcdefg | – | – | |
| Middle atlas | Ofi-351 | 31.00 +8.13 abcd | 30.13 +10.01 cdefghi | 1.75 +0.50 ef | 21.07 +2.08 defghijkl | 11.93 +0.99 bcd | 0.56 +0.01 bcdef | 0.77 +0.29 abc | 3.03 +0.48 bcdefg | – | – | |
| Northeast Morocco | Ofi-421 | 31.00 +7.43 abcd | 32.68 +9.46 cdefghi | 1.50 +0.58 ef | 23.48 +1.07 cdefghijkl | 11.53 +1.02 bcd | 0.52 +0.06 bcdef | 0.73 +0.21 abc | 2.45 +0.26defg | – | – | |
| Northeast Morocco | Ofi-451 | 24.50 +1.78 bcd | 39.63 +12.96 abcdefghi | 2.50 +0.58 cdef | 23.60 +1.31 cdefghijkl | 11.30 +0.41 bcd | 0.47 +0.01 bcdef | 0.70 +0.00 abc | 2.40 +0.08 efg | – | – | |
| Eastern Rif | Ofi-521 | 21.88 +6.51 cd | 21.50 +10.01 hi | 1.00 +0.00f | 17.00 +1.63 jkl | 12.00 +1.63 bcd | 0.70 +0.16 bc | 1.10 +0.08 abc | 3.50 +0.41 abcde | – | – | |
| Eastern Rif | Ofi-531 | 21.33 +6.55 cd | 23.33 +7.32 ghi | 1.00 +0.00f | 22.50 +0.41 cdefghijkl | 12.00 +1.63 bcd | 0.53 +0.02 bcdef | 1.00 +0.16 abc | 2.60 +0.33 cdefg | – | – | |
| Eastern Rif | Ofi-533 | 32.40 +14.02 abcd | 42.30 +23.93 abcdefghi | 3.75 +2.36 bcdef | 26.93 +1.37 bcdefghij | 12.90 +0.51 bc | 0.48 +0.04 bcdef | 0.73 +0.21 abc | 4.17 +1.31a | – | – | |
| Eastern Rif | Ofi-542 | 26.38 +15.21 abcd | 29.75 +21.63 defghi | 2.00 +0.82def | 29.05 +1.02 bcdef | 14.85 +0.78 bc | 0.51 +0.01 bcdef | 0.85 +0.04 abc | 3.70 +0.08abc | – | – | |
| Eastern Rif | Ofi-561 | 37.25 +5.84 abcd | 34.93 +14.14 bcdefghi | 1.75 +0.96 ef | 26.20 +6.49 bcdefghijk | 12.75 +3.52 bc | 0.48 +0.05 bcdef | 0.78 +0.30 abc | 2.70 +0.48 bcdefg | – | – | |
| Western Rif | Ofi-612 | 36.25 +8.87 abcd | 33.63 +9.32 cdefghi | 2.75 +0.50 cdef | 23.10 +2.21 cdefghijkl | 11.57 +1.05 bcd | 0.50 +0.03 bcdef | 1.03 +0.09 abc | 2.37 +0.45 efg | – | – | |
| Western Rif | Ofi-631 | 30.20 +9.41 abcd | 33.88 +13.14 cdefghi | 3.50 +0.58 bcdef | 21.00 + 0.41 defghijkl | 12.25 +0.20 bcd | 0.58 +0.00 bcdef | 0.75 +0.04 abc | 2.80 +0.00 bcdefg | – | – | |
| Agadir | Ofi-711 | 41.70 +5.77 abcd | 50.80 +9.72 abcdefghi | 2.75 +1.26 cdef | 28.38 +2.84 bcdefg | 12.90 +2.37 bc | 0.49 +0.15 bcdef | 0.55 +0.19bc | 3.03 +0.90 bcdefg | – | – | |
| Agadir | Ofi-712 | 40.17 +6.54 abcd | 57.50 +20.00 abcdefgh | 4.25 +2.63 bcdef | 26.03 +2.53 bcdefghijk | 15.47 +2.17 bc | 0.56 +0.04 bcdef | 0.87 +0.17 abc | 2.70 +0.22 bcdefg | – | – | |
| Agadir | Ofi-713 | 45.43 +4.64 ab | 38.75 +8.21 abcdefghi | 2.25 +1.26def | 34.00 +3.58ab | 20.23 +10.73a | 0.58 +0.25 bcdef | 0.83 +0.26 abc | 3.00 +0.08 bcdefg | – | – | |
| Agadir | Ofi-714 | 35.78 +6.63 abcd | 47.43 +8.25 abcdefghi | 2.50 +0.58 cdef | 25.15 +9.11 bcdefghijk | 15.78 +3.26 bc | 0.67 +0.21 bcd | 0.86 + 0.21 abc | 2.85 +0.24 bcdefg | – | – | |
| Agadir | Ofi-723 | 39.03+11.53 abcd | 43.88 +15.29 abcdefghi | 2.25 +0.96def | 28.45 +2.19 bcdefg | 15.08 +0.87 bc | 0.53 +0.04 bcdef | 1.00 +0.16 abc | 3.05 +0.10 bcdefg | – | – | |
| Agadir | Ofi-731 | 36.85 +4.98 abcd | 48.38 +7.62 abcdefghi | 2.00 +0.82def | 27.43 +1.54 bcdefghij | 14.13 +1.26 bc | 0.51 +0.03 bcdef | 0.88 +0.28 abc | 2.95 +0.54 bcdefg | – | – | |
| Agadir | Ofi-732 | 42.68 +8.01 abc | 40.58 +8.39 abcdefghi | 2.50 +0.58 cdef | 29.00 +3.24 bcdef | 14.80 +0.99 bc | 0.51 +0.03 bcdef | 1.10 +0.22 abc | 3.13 +0.25 bcdefg | – | – | |
| Agadir | Ofi-733 | 38.25 + 10.44 abcd | 42.65 +16.01 abcdefghi | 2.25 +0.50def | 27.65 +8.00 bcdefghi | 13.48 +2.67 bc | 0.48 +0.05 bcdef | 1.13 +0.56 abc | 3.00 +0.28 bcdefg | – | – | |
| Agadir | Ofi-742 | 42.93 +9.94 abc | 56.25 +23.09 abcdefgh | 3.75 +1.71 bcdef | 27.00 +2.27 bcdefghij | 13.33 +1.68 bc | 0.49 +0.07 bcdef | 0.75 +0.24 abc | 2.98 +0.24 bcdefg | – | – | |
| Agadir | Ofi-743 | 39.75 +10.43 abcd | 69.88 +26.42 ab | 4.25 +3.30 bcdef | 25.98 +4.29 bcdefghijk | 12.95 +1.32 bc | 0.50 +0.06 bcdef | 0.63 +0.29 abc | 3.03 +0.40 bcdefg | – | – | |
| Agadir | Ofi-751 | 45.75 +2.72 ab | 50.88 +19.84 abcdefghi | 4.00 +1.63 bcdef | 32.68 +10.79 abc | 13.13 +1.11 bc | 0.44 +0.18 cdef | 1.00 +0.26 abc | 3.05 +0.33 bcdefg | – | – | |
| Agadir | Ofi-752 | 46.18 +3.94 ab | 64.63 +6.42 abcde | 5.25 +0.50bcd | 31.15 +2.56 abcd | 15.63 +0.75 bc | 0.50 +0.06 bcdef | 0.73 +0.24 abc | 2.88 +0.48 bcdefg | – | – | |
| Agadir | Ofi-761 | 40.00 +10.58 abcd | 55.38 +19.74 abcdefgh | 3.50 +1.29 bcdef | 22.45 +6.94 cdefghijkl | 13.43 +3.06 bc | 0.63 +0.22bcde | 0.73 +0.40 abc | 2.68 +0.24 bcdefg | – | – | |
| Agadir | Ofi-762 | 48.25 + 5.12 a | 45.00 +13.20 abcdefghi | 3.00 +1.41 cdef | 23.65 +3.49 cdefghijkl | 13.25 +1.46 bc | 0.56 +0.08 bcdef | 0.88 +0.34 abc | 2.68 +0.30 bcdefg | – | – | |
| Agadir | Ofi-771 | 33.38 +3.25 abcd | 46.00 +5.07 abcdefghi | 3.00 +1.41 cdef | 25.83 +1.72 bcdefghijk | 12.05 +1.46 bcd | 0.46 +0.05 bcdef | 0.90 +0.16 abc | 2.75 +0.65 bcdefg | – | – | |
| Agadir | Ofi-772 | 33.93 +3.00 abcd | 47.07 +12.40 abcdefghi | 2.00 +0.82def | 25.73 +1.46 bcdefghijk | 12.80 +1.12 bc | 0.49 +0.05 bcdef | 0.73 +0.25 abc | 2.63 +0.19 bcdefg | – | – | |
| O. megacantha | Jamaât Riah | Om-A1 | 30.43 +3.54 abcd | 42.05 +15.92 abcdefghi | 2.75 +0.96 cdef | 25.38 +3.50 bcdefghijk | 12.38 +0.48 bcd | 0.49 +0.07 bcdef | 0.80 +0.18 abc | 2.68 +0.39 bcdefg | 4.25 + 1.50 abc | 2.70 + 0.22ghijklmno |
| Jamaât Riah | Om-A2 | 32.15 + 2.83 abcd | 43.05 +13.93 abcdefghi | 2.00 +0.00def | 25.15 +1.65 bcdefghijk | 14.67 +1.31 bc | 0.58 +0.03 bcdef | 0.90 +0.16 abc | 2.90 +0.16 bcdefg | 3.75 +0.50cd | 3.00 +0.50 cdefghijklm | |
| Jamaât Riah | Om-A3 | 29.03 +5.85 abcd | 36.60 +6.13 bcdefghi | 2.75 +0.96 cdef | 25.40 +1.55 bcdefghijk | 12.80 +0.98 bc | 0.50 +0.01 bcdef | 1.05 +0.04 abc | 3.10 +0.33 bcdefg | 3.00 +0.00d | 2.15 +0.29 nop | |
| Jamaât Riah | Om-B1 | 29.03 +5.85 abcd | 36.60 +6.13 bcdefghi | 1.75 +0.50 ef | 25.25 +1.56 bcdefghijk | 12.35 +1.11 bcd | 0.49 +0.02 bcdef | 0.98 +0.10 abc | 2.95 + 0.37 bcdefg | 3.00 + 0.00d | 2.38 +0.39 jklmno | |
| Jamaât Riah | Om-K1 | 25.00 +10.39 bcd | 21.65 +12.88 hi | 1.00 +0.00f | 23.53 +0.38 cdefghijkl | 13.00 +1.63 bc | 0.55 +0.04 bcdef | 1.30 +0.08 abc | 3.00+0.41 bcdefg | 4.00 +0.82 bcd | 2.10 +0.08 op | |
| Jamaât Riah | Om-K2 | 33.93 +9.72 abcd | 41.88 +10.36 abcdefghi | 3.50 +1.00 bcdef | 26.30 +3.50 bcdefghijk | 12.83 +2.42 bc | 0.48 +0.03ef | 1.40+1.33a | 2.90 +0.36 bcdefg | 3.00 +0.82 d | 3.07 +0.51 cdefghijk | |
| Jamaât Riah | Om-K3 | 32.63 +7.87 abcd | 65.38 +14.10 abcd | 4.25 +0.96 bcdef | 26.28 +1.21 bcdefghijk | 14.05 +1.17 bc | 0.53 +0.03 bcdef | 0.68 +0.33 abc | 3.05 +0.17 bcdefg | 3.50 +0.58 cd | 2.60 +0.66hijklmno | |
| Rhamna | Om-111 | 29.50 + 3.27 abcd | 50.05 +3.23 abcdefghi | 2.50 +0.58 cdef | 24.85 +1.51 bcdefghijk | 11.00 +0.00 bcd | 0.44 +0.02 cdef | 0.60 +0.00 abc | 2.85 +0.12 bcdefg | 4.00 +0.00bcd | 2.70 +0.00ghijklmno | |
| Rhamna | Om-112 | 26.38 +2.06 abcd | 30.13 +7.88 cdefghi | 1.50 +0.58 ef | 19.83 +4.19 fghijkl | 9.40 +1.04cd | 0.48 +0.06 bcdef | 0.53 +0.05c | 2.67 +0.47 bcdefg | 3.00 +0.00 d | 2.27 +0.12 mnop | |
| Rhamna | Om-122 | 28.90 +5.32 abcd | 42.58 +8.63 abcdefghi | 3.00 +1.41 cdef | 23.17 +2.05 cdefghijkl | 14.33 +3.52 bc | 0.51 +0.01 bcdef | 0.80 +0.33 abc | 2.70 +0.50 bcdefg | 3.00 +0.00 d | 2.77 +0.46ghijklmno | |
| Rhamna | Om-131 | 37.75 +3.88 abcd | 55.38 +20.27 abcdefgh | 3.75 +2.36 bcdef | 23.75 +4.17 cdefghijkl | 13.30 +2.03 bc | 0.56 +0.03 bcdef | 1.00 +0.47 abc | 3.03 +0.67 bcdefg | 4.00 +0.00 bcd | 3.80 +0.58 ab | |
| Rhamna | Om-143 | 24.67 +0.62 bcd | 34.07 +3.35 cdefghi | 2.50 +1.73 cdef | 22.70 +3.84 cdefghijkl | 9.75 +0.20cd | 0.45 +0.09 bcdef | 0.70 +0.16 abc | 2.55 +0.04 cdefg | 3.50 +0.58 cd | 2.30 +0.08 lmnop | |
| Rhamna | Om-181 | 27.28 +4.30 abcd | 32.88 +5.42 cdefghi | 1.00 +0.00f | 37.00 +1.63a | 12.50 +0.41 bcd | 0.33 +0.02f | 0.90 +0.08 abc | 3.00 +0.08 bcdefg | 4.00 +0.82 bcd | 1.70 +0.16 p | |
| Ouardigha | Om-221 | 25.13 +8.13 bcd | 39.25 +7.71 abcdefghi | 2.00 +0.82def | 20.63 +1.25 defghijkl | 10.88 +0.75 bcd | 0.52 +0.03 bcdef | 0.75 +0.26 abc | 2.38 +0.15 efg | 3.50 +0.58 cd | 2.98 +0.66defghijklm | |
| Ouardigha | Om-251 | 29.03 + 4.45 abcd | 37.75 +11.26 abcdefghi | 2.50 +0.58 cdef | 21.70 +0.73 defghijkl | 12.50 +0.82 bcd | 0.57 +0.02 bcdef | 1.00 +0.08 abc | 3.00 +0.00 bcdefg | 3.00 +0.00 d | 2.30 +0.00 lmnop | |
| Middle atlas | Om-267 | 31.63 +2.17 abcd | 43.38 +8.36 abcdefghi | 2.75 +2.36 cdef | 20.93 +2.81 defghijkl | 11.15 +1.35 bcd | 0.53 +0.04 bcdef | 0.70 +0.18 abc | 2.60 + 0.34cdefg | 3.50 +1.00 cd | 2.75 +0.80ghijklmno | |
| Middle atlas | Om-352 | 31.88 +6.76 abcd | 42.15 +13.78 abcdefghi | 2.75 +0.50 cdef | 22.83 +2.25 cdefghijkl | 12.70 +0.92 bc | 0.55 +0.02 bcdef | 0.77 +0.26 abc | 2.90 +0.43 bcdefg | 3.50 +1.00 cd | 2.83 +0.40efghijklmn | |
| Middle atlas | Om-371 | 32.50 +6.54 abcd | 42.38 +12.26 abcdefghi | 2.25 +1.26def | 25.50 +3.97 cdefghijkl | 12.75 +1.85 bc | 0.57 +0.18 bcdef | 0.83 +0.15 abc | 2.75 +0.33 bcdefg | 5.00 +1.15a | 3.08 +0.49 cdefghijk | |
| Northeast Morocco | Om-411 | 39.73 +8.94 abcd | 58.50 +7.93 abcdefg | 3.25 +0.50 cdef | 25.30 +2.89 bcdefghijk | 13.95 +2.42 bc | 0.55 +0.05 bcdef | 0.73 +0.24 abc | 3.15 +0.83 bcdefg | 4.25 +0.50 abc | 3.13 +0.57 cdefghi | |
| Northeast Morocco | Om-422 | 30.63 +2.02 abcd | 44.63 +7.38 abcdefghi | 2.50 +1.73cdef | 22.88 +1.11 cdefghijkl | 11.85 +2.28 bcd | 0.52 +0.07 bcdef | 0.80 +0.32 abc | 2.73 +0.25 bcdefg | 3.75 +0.50 cd | 3.35 +0.13bcdefg | |
| Northeast Morocco | Om-431 | 38.85 +7.35 abcd | 40.95 +19.31 abcdefghi | 3.00 +0.82 cdef | 23.67 +1.43 cdefghijkl | 13.23 +0.76 bc | 0.55 +0.03 bcdef | 0.80 +0.22 abc | 2.75 +0.21 bcdefg | 4.50 +0.58 abc | 3.57 +0.39 abcd | |
| Northeast Morocco | Om-441 | 36.63 + 7.23 abcd | 43.35 +10.61 abcdefghi | 1.75 +0.96 ef | 26.25 +4.52 bcdefghijk | 12.97 +0.54 bc | 0.47 +0.06 bcdef | 1.20 +0.85 abc | 2.63 +0.26 bcdefg | 3.00 +0.00 d | 2.97 +0.12defghijklm | |
| Northeast Morocco | Om-442 | 35.25 +3.57 abcd | 54.50 +7.33 abcdefgh | 2.00 +0.00def | 26.43 +2.32 bcdefghijk | 13.60 +0.90 bc | 0.51 +0.01 bcdef | 0.75 +0.21 abc | 2.85 +0.39 bcdefg | 3.75 +0.50 cd | 3.28 +1.03 bcdefgh | |
| Northeast Morocco | Om-461 | 29.80 +12.09 abcd | 34.15 +15.09 cdefghi | 2.50 +0.58 cdef | 22.55 +2.59 cdefghijkl | 12.40 +1.73 bcd | 0.54 +0.02 bcdef | 0.87 +0.17 abc | 2.43 +0.31 efg | 3.50 +0.58 cd | 2.53 +0.34 ijklmno | |
| Northeast Morocco | Om-471 | 35.00 +5.72 abcd | 33.00 +8.16 cdefghi | 1.00 +0.00f | 23.75 +1.84 cdefghijkl | 13.75 +1.02 bc | 0.57 +0.01 bcdef | 0.85 +0.04 abc | 3.05 +0.04 bcdefg | 3.50 +0.58 cd | 2.80 +0.08fghijklmno | |
| Eastern Rif | Om-511 | 28.13 +8.93 abcd | 30.20 +10.41 cdefghi | 3.50 +0.58 bcdef | 24.25 +3.06 bcdefghijk | 12.90 + 1.96 bc | 0.52 +0.01 bcdef | 1.10 +0.00 abc | 2.50 +0.00 defg | 4.00 +0.00 bcd | 2.85 +0.53efghijklmn | |
| Eastern Rif | Om-532 | 36.50 +6.16 abcd | 45.40 +20.76 abcdefghi | 3.00 +1.15 cdef | 20.98 +3.81 defghijkl | 12.40 +2.09 bcd | 0.59 + 0.05 bcdef | 0.80 +0.20 abc | 2.75 +0.21 bcdefg | 4.00 +0.00 bcd | 2.43 +0.22 ijklmno | |
| Eastern Rif | Om-541 | 37.38 +6.14 abcd | 40.75 +15.02 abcdefghi | 2.00 +0.82def | 22.75 +3.84 cdefghijkl | 12.38 +1.60 bcd | 0.55 +0.06 bcdef | 0.80 +0.29 abc | 2.73 +0.46 bcdefg | 3.50 +0.58 cd | 2.43 +0.49 ijklmno | |
| Eastern Rif | Om-543 | 39.13 + 6.14 abcd | 38.58 +9.14 abcdefghi | 2.25 +0.50def | 20.90 +2.31 defghijkl | 11.03 +1.86 bcd | 0.52 +0.06 bcdef | 0.85 +0.13 abc | 2.13 +0.25 g | 3.75 +0.50 cd | 2.88 +0.25efghijklmn | |
| Eastern Rif | Om-551 | 31.75 +5.39 abcd | 44.50 +7.82 abcdefghi | 2.50 +1.73 cdef | 20.50 +4.02 efghijkl | 13.85 +7.67 bc | 0.70 +0.19bc | 0.90 +0.16 abc | 2.30 +0.38 fg | 4.00 +0.00 bcd | 2.35 +0.64 klmno | |
| Eastern Rif | Om-571 | 40.33 +5.37 abcd | 35.50 +10.45 bcdefghi | 2.50 +1.73 cdef | 23.80 +2.03 cdefghijkl | 13.23 +1.34 bc | 0.55 +0.04 bcdef | 0.95 +0.10 abc | 2.33 +0.15 fg | 4.25 +0.50 abc | 2.40 +0.12 ijklmno | |
| Eastern Rif | Om-572 | 37.50 + 11.02 abcd | 35.00 +4.49 bcdefghi | 1.00 +0.00f | 25.35 +4.20 bcdefghijk | 13.85 +3.55 bc | 0.53 +0.05 bcdef | 1.05 +0.20 abc | 2.30 +0.41 fg | 3.50 +0.58 cd | 2.30 +0.08 lmnop | |
| Eastern Rif | Om-581 | 35.63 +9.07 abcd | 40.38 +7.80 abcdefghi | 4.00 +1.83 bcdef | 21.13 +1.18 defghijkl | 11.40 +0.86 bcd | 0.54 +0.03 bcdef | 0.85 +0.06 abc | 2.05 +0.31 g | 4.25 +0.50 abc | 2.85 +0.10efghijklmn | |
| Western Rif | Om-632 | 37.45 +4.59 abcd | 30.68 +7.81 cdefghi | 3.00 +0.82 cdef | 17.30 +4.87 ijkl | 11.50 +1.35 bcd | 0.55 +0.06 bcdef | 0.70 +0.20 abc | 2.48 +0.13 defg | 4.00 +0.00 bcd | 2.30 +0.14 lmnop | |
| Western Rif | Om-621 | 29.00 +7.82 abcd | 28.13 +7.20 fghi | 2.50 +1.29 cdef | 21.75 +0.20 defghijkl | 11.00 +0.00 bcd | 0.65 +0.12 bcd | 0.90 +0.08 abc | 2.35 +0.29 efg | 3.50 +0.58 cd | 2.35 +0.12 klmno | |
| Western Rif | Om-651 | 38.13 + 7.40 abcd | 54.00 +6.87 abcdefgh | 2.75 +0.50 cdef | 22.93 +3.20 cdefghijkl | 13.40 +2.09 bc | 0.58 +0.02 bcdef | 0.80 +0.22 abc | 2.43 +0.38 efg | 4.00 +0.00 bcd | 2.95 +0.50defghijklm | |
| Agadir | Om-722 | 31.98 + 4.04 abcd | 41.50 +8.50 abcdefghi | 3.50 +0.58 bcdef | 24.23 +1.93 bcdefghijk | 12.56 +1.70 bcd | 0.51 +0.03 bcdef | 0.80 +0.24 abc | 2.63 +0.26 bcdefg | 4.00 +0.00 bcd | 3.03 +0.05 cdefghijkl | |
| Agadir | Om-741 | 31.63 +3.12 abcd | 57.43 +6.95 abcdefgh | 3.75 +1.26 bcdef | 23.75 +1.44 cdefghijkl | 11.95 +1.14 bcd | 0.50 +0.04 bcdef | 0.85 +0.47 abc | 2.08 +0.61 g | 3.75 +0.50 cd | 2.63 +0.62 hijklmno | |
| O. robusta | Northeast Morocco | Or-412 | 29.28 +6.52 abcd | 42.20 +6.37 abcdefghi | 2.75 +0.96 cdef | 17.90 +3.59 ghijkl | 17.33 +3.45ab | 0.97 +0.10a | 1.38 +0.05ab | 3.60 +0.45abcd | 4.50 +0.58 abc | 4.03 +0.05a |
| O. aequatorialis | Rhamna | Oa-171 | 39.88 +6.94 abcd | 34.63 +4.70 cdefghi | 3.25 +0.50 cdef | 30.63 +2.95 abcde | 14.03 +9.99 bc | 0.48 +0.41 bcdef | 1.08 +0.39 abc | 2.43 +0.33 efg | 5.00 +0.82 a | 3.48 +0.32 abcdef |
| O. dillenii | Ouardigha | Od-242 | 19.85 +2.16 d | 28.50 +3.67 efghi | 7.75 +3.50a | 13.75 +1.84l | 6.25 +0.86d | 0.45 +0.00 bcdef | 0.65 +0.04 abc | 2.90 +0.16 bcdefg | – | – |
| O. leucotricha | Middle atlas | Ol-361 | 37.63 +3.35 abcd | 53.33 +9.51 abcdefgh | 4.00 +0.82 bcdef | 24.75 + 3.10 bcdefghijk | 14.38 +0.25 bc | 0.66 +0.19 bcd | 1.13 +0.31 abc | 2.70 +0.26 bcdefg | 5.00 +0.82 a | 3.85 +0.54 ab |
| Western Rif | Ol-633 | 42.55 +9.17 abc | 42.93 +18.69 abcdefghi | 2.25 +1.26def | 24.40 +5.63 bcdefghijk | 13.18 +2.19 bc | 0.55 +0.10 bcdef | 0.93 +0.32 abc | 2.90 +0.12 bcdefg | 4.50 +1.29 abc | 3.53 +1.01 abcde | |
| Western Rif | Ol-611 | 45.88 +1.11 ab | 50.50 +6.81 abcdefghi | 4.00 +1.63 bcdef | 27.30 +3.50 bcdefghij | 13.63 +1.49 bc | 0.50 +0.02 bcdef | 0.85 +0.24 abc | 2.73 +0.38 bcdefg | 4.25 +0.50 abc | 3.65 +0.84 abc | |
| Western Rif | Ol-641 | 40.98 +8.04 abcd | 51.95 +13.07 abcdefghi | 3.25 +0.50 cdef | 23.95 +5.17 cdefghijk | 13.45 +2.09 bc | 0.54 +0.03 bcdef | 0.85 +0.24 abc | 2.62 +0.26 bcdefg | 4.75 +0.96 ab | 3.10 +1.05 cdefghij | |
| O. inermis | Rhamna | Oi-191 | 35.63 +8.94 abcd | 50.00 +8.99 abcdefghi | 5.75 +0.96bc | 20.73 +3.26 defghijkl | 9.60 +1.82cd | 0.46 +0.02 bcdef | 0.78 +0.28 abc | 2.33 +0.25 fg | – | – |
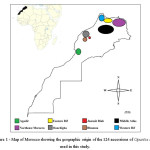 |
Figure 1: Map of Morocco showing the geographic origin of the 124 accessions of Opuntia spp. used in this study. Click here to View figure |
Molecular Analysis
DNA Extraction
In this study, we used 22 ecotypes from different geographic origins in Morocco, and which represent seven species of Opuntia identified from the morphological traits; for example, for O. ficus indica, we used 8 accessions, each one was collected from a different region. Total genomic DNA was extracted according to the protocol of Doyle and Doyle (1990)17 with some slight modifications, using 1 g of lyophilized and ground cladode tissue. The DNA concentration was then diluted to 20 ng/µL with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) using a biophotometer (Eppendorf).
ISSR Analysis
Fourteen ISSR primers (Table 2) purchased either from Operon Technologies Inc. (Alameda, CA, USA) or Qiagen (Valencia, CA, USA) were tested in this study; and 2.5 µL of each primer was added to a PCR mixture containing 3 µL DNA template, 12.3 µL sterile distilled water, 3.5 µL (25 mM) MgCl2, 1 µL(10 mM) dNTPs, 0.2 µL(5u/µL) Taq polymerase and 2.5 µL Taq buffer (All purchased from Promega, Madison, WI, USA). DNA amplifications were performed using a DNA thermal cycler (Techne) with the following parameters: initial denaturation at 94°C for 5 min, 45 cycles of 15 s of denaturation at 94 °C, 30 s of annealing at the specific temperature of each primer (Table 2), and 60 s of extension at 72°C; then a final extension at 72 °C for 7 min.
Table 2: List and characteristics of primers used to assess genetic variability within and among Opuntia spp in Morocco.
|
Primer type |
Primer code | Primer sequence 5’- 3’ | Annealing temperature (°C) | Total number of bands | Number of polymorphic bands | Sizerange(bp) |
PIC |
| ISSR | ISSR-8 | ATCAATCAATCAATCA | 30 | 7 | 7 | 234-1966 | 0.97 |
| ISSR-17 | ATCATCATCATCATC | 54 | 4 | 4 | 225-1268 | 0.93 | |
| ISSR-29 | TGTATGTATGTATGTATGTACG | 54 | 6 | 6 | 258-731 | 0.62 | |
| ISSR-31 | GACAGACAGACAGACAGACA | 54 | 9 | 9 | 125-1047 | 0.90 | |
| ISSR-34 | CCTGGGTTCC | 71 | 7 | 7 | 920-2077 | 0.86 | |
| ISSR-38 | GGTGGCGGGA | 71 | 8 | 7 | 391-1275 | 0.80 | |
| ISSR-42 | ATCATCATCATCATCATCATC | 60 | – | – | – | – | |
| ISSR-46 | ATCATCATCATCATC | 60 | 8 | 7 | 158-2127 | 0.84 | |
| ISSR-48 | CATCAGAAGTGCGTTCGTAGG | 60 | 3 | 3 | 400-900 | 0.78 | |
| ISSR-49 | TGTGTGTGTGTGTGTGTGTG | 60 | – | – | – | – | |
| ISSR-50 | AGGAGGAGGAGGAGGAGGAGGAGGTC | 73 | 7 | 7 | 271-889 | 0.80 | |
| ISSR-51 | TCCTCCTCCTCCTCCAG | 60 | – | – | – | – | |
| ISSR-53 | TCTCTCTCTCTCTCTCTCTC | 71 | – | – | – | – | |
| ISSR-54 | TCCTCCTCCTCCTCCG | 52 | 7 | 7 | 191-1047 | 0.77 | |
| Total | 14 | 66 | 64 | ||||
| Mean | 6.6 | 6.4 | 0.82 | ||||
| RAPD | UBC-515 | GGGGGCCTCA | 36 | – | – | – | – |
| UBC-518 | TGCTGGTCCA | 36 | – | – | – | – | |
| UBC-523 | ACAGGCAGAC | 36 | – | – | – | – | |
| UBC-528 | GGATCTATGC | 36 | – | – | – | – | |
| UBC-529 | CACTCCTACA | 36 | – | – | – | – | |
| UBC-531 | GCTCACTGTT | 36 | – | – | – | – | |
| UBC-533 | GCATCTACGC | 36 | – | – | – | – | |
| UBC-536 | GCCCCTCGTC | 36 | – | – | – | – | |
| UBC-547 | TATGACCTGG | 36 | – | – | – | – | |
| UBC-548 | GTACATGGGC | 36 | – | – | – | – | |
| UBC-551 | GGAAGTCCAC | 36 | – | – | – | – | |
| UBC-562 | CAAAGTAGCC | 36 | – | – | – | – | |
| UBC-565 | GGTCGATTTC | 36 | – | – | – | – | |
| UBC-566 | CCACATGCGA | 36 | – | – | – | – | |
| UBC-567 | AGACACCTGA | 36 | – | – | – | – | |
| UBC-568 | ACCTGTTCTC | 36 | – | – | – | – | |
| UBC-569 | CGAATTGCTG | 36 | – | – | – | – | |
| UBC-570 | GGCCGCTAAT | 36 | – | – | – | – | |
| UBC-571 | GCGCGGCACT | 36 | 6 | 6 | 464-1375 | 0.76 | |
| UBC-572 | TTCGACCATC | 36 | – | – | – | – | |
| UBC-573 | CCCTAATCAG | 36 | – | – | – | – | |
| UBC-574 | GCCAGACAAG | 36 | – | – | – | – | |
| UBC-577 | GTCTGATGTG | 36 | – | – | – | – | |
| UBC-584 | GCGGGCAGGA | 36 | – | – | – | – | |
| UBC-587 | GCTACTAACC | 36 | – | – | – | – | |
| UBC-588 | CAGAGGTTGG | 36 | – | – | – | – | |
| UBC-589 | GACGGAGGTC | 36 | – | – | – | – | |
| UBC-591 | TCCCTCGTGG | 36 | – | – | – | – | |
| UBC-592 | GGGCGAGTGC | 36 | – | – | – | – | |
| UBC-593 | CGAGCTTTGA | 36 | – | – | – | – | |
| UBC-596 | CCCCTCGAAT | 36 | – | – | – | – | |
| UBC-598 | ACGGGCGCTC | 36 | – | – | – | – | |
| UBC-599 | CAAGAACCGC | 36 | – | – | – | – | |
| UBC-600 | GAAGAACCGC | 36 | – | – | – | – | |
| Total | 34 | 6 | 6 | ||||
| Mean | 6 | 6 | 0.76 |
RAPD Analysis
For RAPD analysis, 34 RAPD primers (Table 2) purchased from The University of British Columbia (UBC; Vancouver, Canada) were tested. The PCR mixture was containing the same above-mentioned components for ISSR primers, but with substituting ISSR primers with 0.4 µM of each RAPD primer. Amplification conditions were as follows: initial denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94 °C for 60 s, annealing at 36 °C for 60 s, and extension at 72 °C for 90 s; with a final extension at 72 °C for 15 min.
ISSR and RAPD Gel Electrophoresis
After amplification, 5 µL bromophenol blue (Sigma, St. Louis, MO, USA) was added to each amplification product. Afterwards, 8 µL of each sample was electrophoresed at 90 V, 5 W for 1 h on 1.8 % agarose gel (Promega, Madison, WI, USA) in 100 mL TBE buffer (90 mL sterile distilled water, 10 mL TBE buffer 10X; Promega, Madison, WI, USA) supplemented with 6.6 µL ethidium bromide (Sigma, St. Louis, MO, USA). Molecular weight of the amplified bands was estimated by comparison with lambda DNA/EcoR I and Hind III markers (Promega, Madison, WI, USA) as standard. The samples were then visualized and photographed with a gel documentation system (Fisher Scientific).
Statistical Analysis of Molecular Data
For each gel, we recorded the presence (1) or absence (0) of bands in individual lanes; consequently, a binary data matrix was created. For each marker, we calculated the polymorphic information content (PIC) using the formula developed by Botstein et al. (1980) [7]: PIG = 1-Σnj=1pi2-2Σn1-i=1Σnj= i+1pi2pj2. where pi and pj are the frequencies of the ith and jth alleles and n is the total number of such alleles. The Dice’s coefficients were calculated and a dendrogram was generated based on the similarity matrix and the SAHN module using unweighted pair-group method of arithmetic analysis (UPGMA). Afterwards, a principal component analysis (PCA) was performed. All these analyses were conducted using NTSYSpc v 2.1 software.34
Results and Discussion
Morphological traits to assess genetic variability
Genetic variability within Opuntia ficus indica
Our investigation showed that Opuntia ficus indica is present in all the studied areas. However, our results demonstrated clear differences among the 78 accessions with regard to morphological descriptors. For example, the plant height was ranging from 21.33 cm in Ofi-531 (from Eastern Rif) to 48.25 cm in Ofi-762 (from Agadir) and the plant diameter was ranging from 16.43 cm in Ofi-275 (from Ouardigha) to 71.38 in Ofi-163 (from Rhamna). In some cases, accessions from the same geographical site exhibited significantly different measures of morphological traits; for example, in accessions from Rhamna, cladode length was ranging from 16.30 cm in Ofi-141 to 29.13 cm in Ofi-163 (Table 1). The O. ficus indica accessions showed similarity coefficients ranging from 0.00 to 1.00 (proximity matrix not shown). The lowest similarity coefficient was noticed between accessions Ofi-212 and Ofi-262 from Ouardigha and between accessions Ofi-771 and Ofi-772 from Agadir while the highest similarity coefficient was observed between accessions Ofi-163 (from Rhamna) and Ofi-275 (from Ouardigha). In some cases, accessions from different regions show very low similarity coefficient (e.g. Ofi-C3 from Jamaât Riah and Ofi-561 from Eastern Rif displayed a similarity coefficient of 0.001). The dendrogram presented in Figure 2 suggested 3 major clusters. The first cluster contains 30 accessions (38.46 % of the total accessions); the second cluster contains 5 accessions (6.41 % of the total accessions) and the third cluster is comprised of 43 accessions (55.13% of the total accessions). Surprisingly, the three clusters contain accessions collected from different geographical sites.
 |
Figure 2: Dendrogram grouping 78 accessions of Opuntia ficus indica based on 10 morphological traits and Ward’s method. The red stars indicate accessions used for molecular analysis. Click here to View figure |
Genetic variability within Opuntia megacantha
The plant height of O. megacantha accessions ranged from 24.67 cm to 40.33 cm, plant diameter from 21.65 cm to 65.38 cm, the average number of cladodes per plant from 1.00 to 4.25, cladode height from 17.30 cm to 37.00 cm, cladode width from 9.40 cm to 14.67 cm, the index of cladode shape from 0.33 to 0.70, cladode thickness from 0.53 cm to 1.40 cm, the mean distance between areoles from 2.05 cm to 3.15 cm, the average number of spines per areole from 3.00 to 5.00 and the mean length of the longest spine per areole from 1.70 cm to 3.80 cm. Here again, some accessions from the same region displayed significantly different measures of morphological traits (Table 1). The range of similarity coefficients was from 0.00 to 1.00 (proximity matrix not shown). The lowest similarity was between Om-B1 and Om-A3, both from Jamaât Riah, while the highest similarity was between Om-K1 and Om-K3, from Jamaât Riah too. The dendrogram (Figure 3) shows 3 clusters, the first cluster consisted of 11 accessions (28.95 % of the total accessions); the second cluster contains 20 accessions (52.63 % of the total accessions) whereas the third cluster comprised 7 accessions (18.42 % of the total accessions). Interestingly, accessions collected from different regions could be found within the same cluster.
 |
Figure 3: Dendrogram grouping 38 accessions of Opuntia megacantha based on 10 morphological traits and Ward’s method. The red stars indicate accessions used for molecular analysis. Click here to View figure |
Genetic variability among Opuntia spp
Taking into consideration all the studied accessions of Opuntia spp., the plant height ranged from 19.85 cm in Od-242 to 48.25 cm in Ofi-762, the average number of cladodes per plant from 1.00 to 7.75 in Od-242, with cladodes of different heights (from 13.70 in Ofi-331 to 37.00 in Om-181) and widths (from 6.25 cm in Od-242 to 20.23 in Ofi-713). Cladode thickness varied from 0.47 cm in Ofi-264 to 1.40 cm in Om-K2 (Table 1). Among the studied species, O. ficus indica, O. dillenii and O. inermis are spineless. The similarity coefficients were ranging from 0.00 to 1.00. The dendrogram shows three different clusters (Figure 4), and accessions from different species or geographic origins could be gathered into the same cluster. This shows that the clusters obtained do not fit with the species or geographical sites evaluated.
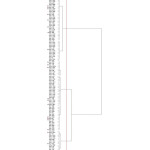 |
Figure 4: Dendrogram grouping 124 accessions of Opuntia spp. based on 10 morphological traits and Ward’s method. The red stars indicate accessions used for molecular analysis. Click here to View figure |
Molecular markers to assess genetic diversity
After screening 14 ISSR and 34 RAPD primers, 10 ISSR and 1 RAPD primers. were selected for producing unambiguous, reproducible and intense bands. The ISSR primers generated a total of 66 bands, 64 (96.9 %) of which were polymorphic. The number of bands observed per locus varied from 4 to 9, with an average of 6.6. The RAPD primer (UBC-571) gave 6 bands, all polymorphic (Table 2). Based on the PIC values, the markers were highly informative. Indeed, the PIC values varied from 0.62 to 0.97, with an average of 0.82. The highest PIC value was observed in primer ISSR-8 while the lowest value was observed in primer ISSR-29 (Table 2). The dendrogram displayed 7 clusters at a similarity of 0.76 (Figure 5). O. inermis was separated from all the other species. The two accessions of O. leucotricha (Ol-361 and Ol-611) were placed into two different groups while accessions Or-412 (O. robusta) and Oa-171 (O. aequatorialis) were gathered into the same cluster. Interestingly, 14 accessions belonging to O. ficus indica and O. megacantha were gathered into the same group. This was confirmed by PCA (Figure 6), which results were similar to those obtained by the dendrogram.
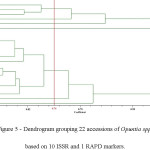 |
Figure 5: Dendrogram grouping 22 accessions of Opuntia spp. based on 10 ISSR and 1 RAPD markers. Click here to View figure |
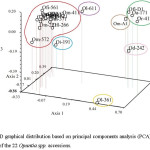 |
Figure 6: 3D graphical distribution based on principal components analysis (PCA) showing the distribution of the 22 Opuntia spp. accessions. Click here to View figure |
Discussion
In the present study, the genetic diversity of 124 accessions of Opuntia spp. was evaluated based on 10 morphological descriptors. The use of morphological descriptors to determine genetic diversity within plant populations is simple, rapid and inexpensive.9 Thus, such descriptors were used in many plant species such as date palm,19 hexaploid oat16 and indigenous rice.43 To the best of our knowledge, this is the first work to assess genetic variability within and among Opuntia populations in Morocco using morphological descriptors. Studies on the genetic variability in Opuntia species using morphological descriptors are scarce. Peña-Valdivia et al. (2008)31 assessed genetic diversity within 55 accessions of Opuntia spp. in Mexico based on 65 morphological descriptors; and indicated that only few morphological traits showed significantly different measures within the studied populations. In a more recent study, Helsen et al. (2009)25 highlighted a clear differentiation between Opuntia species using descriptors based on fruit, flower, seed, spine and trunk characteristics. In our case, the 124 accessions showed significant differences in the measured parameters and were distributed into 3 main distinct clusters based on Ward’s method, containing 56, 45 and 23 accessions, respectively. Interestingly, accessions classified in the same species could be located in different clusters and vice versa, even though some species contain spines while others do not. This may be explained by several factors including the environmental effect and evolutionary change.11-40 Our results are in good agreement with Valadez-Moctezuma et al. (2014)39 who supported the hypothesis about the existence of a smaller number of Opuntia species compared to those currently described, but with high intraspecific genetic variation; and with Labra et al. (2003)27 who suggested that the presence of spines should not be considered as a parameter in Opuntia taxonomy. Our results showed the influence of geographical origins on the morphological traits within the same species. Indeed, in some cases, accessions of the same species but from different regions were gathered into different clusters. This might be due to the environment effect.37-38 Our results confirm previous findings on Opuntia spp. in Sardinia and Corsica islands, which show that environmental factors such as elevation, soil drainage, temperature and rainfall affect plant morphology.21 As far as Opuntia ficus indica is concerned, the use of morphological or agro-morphological traits to assess genetic diversity within its accessions is very scarce. Felker et al. (2005)22 compared genotypes of O. ficus indica from different origins based on yield and fruit characters, and significant differences were observed. In another investigation, Bendhifi et al. (2013)6 compared 28 Tunisian accessions of O. ficus indica using 20 descriptors based on plant, cladode and fruit characteristics. Here again, significant differences were observed between accessions. In our case, the 10 morphological traits showed significant differences between the 78 accessions. Remarkably, some accessions from the same geographical origin displayed significantly different measures of some traits. The use of morphological markers to assess genetic diversity within populations is simple and inexpensive. However, such markers can be affected by the environment.3 Therefore, in the recent years, molecular markers have been widely used to assess genetic diversity within plant populations. Indeed, it was reported that molecular markers cover a larger proportion of the genome than the morphological ones and are less influenced by the environment since genetic variability is evaluated based on genotype rather than phenotype.26-38 In addition, molecular markers have been proven powerful to detect variability when it could not be possible with morphological markers; i.e. a given morphological marker can affect other traits and the usefulness of morphological markers is restricted by their limited number.3 Along this line, molecular markers have been applied alone or in association with morphological descriptors to assess genetic variability in many plant species; e.g. in Phoenix dactylifera L. ,35 Olea europaea L.,8 Citrus sinensis L.29 Concerning the cactus pear, Labra et al. (2003)27 evaluated the genetic diversity among 11 Opuntia species from Italy using chloroplastic simple sequence repeat (cpSSR) and amplified fragment length polymorphism (AFLP) markers, and a high genetic similarity between O. ficus indica and O. megacantha was revealed. Indeed, these authors suggested that O. ficus indica should be considered as a domesticated form of O. megacantha. Caruso et al. (2010)10 assessed the genetic diversity among 62 accessions belonging to 16 Opuntia species from different origins, using simple sequence repeat (SSR) primers, and found that the O. ficus indica accessions did not cluster separately from some Opuntia species, including O. megacantha. In a more recent study, Valadez-Moctezuma et al. (2014)39 assessed the genetic variability among 12 Mexican Opuntia species (52 cultivars) with ISSR and RAPD markers, and reported that O. ficus indica and O. megacantha might have a common ancestry. Our results are consistent with all these authors, since a high genetic similarity was observed between O. ficus indica and O. megacantha. On the other hand, Valadez-Moctezuma et al. (2014)39 screened 120 RAPD primers, of which five were able to generate bands (4.16 %). In our case, only one RAPD primer over 34 (2.94 %) generated bands. Our findings indicated that ISSR markers are the most efficient and informative in Opuntia species. Recently, ISSR markers have been used to assess the genetic and phylogenetic relationships of 15 Opuntia species from Argentina, Bolivia, Brazil, Paraguay, and Uruguay.33 Our findings are in contrast with those of Valadez-Moctezuma et al. (2014),39 who indicated that the assessment of genetic diversity with RAPD markers was relatively more effective than with ISSR markers. In our study, the total number of polymorphic bands (64) and the PIC values (0.62-0.97) of ISSR markers were different from those found by Valadez-Moctezuma et al. (2014),39 in which the number of polymorphic bands was 70 and the PIC varied from 0.21 to 0.27. This difference in polymorphic bands number and PIC values could be explained by the different ISSR primers employed (different sequences), the difference among accessions and the high distance between the geographical sites (Mexico and Morocco). Regarding the species Opuntia ficus indica, Zoghlami et al. (2007)44 used 8 RAPD primers (over 22 screened) to assess the genetic variability within 36 Tunisian accessions, and a considerable genetic diversity was detected. In Morocco, the genetic diversity within O. ficus indica accessions was evaluated using RAPD markers,18 and the effect of the geographical origin was revealed. In our case, findings showed high genetic similarity among the studied accessions of O. ficus indica, even though they were collected from different regions of Morocco.
Conclusion
This paper is the first report to assess the genetic diversity within and among Opuntia species in Morocco using morphological descriptors and molecular markers. A low level of genetic diversity was observed among the accessions analyzed and a high similarity between O. ficus indica and O. megacantha accessions was highlighted. Our findings show also that ISSR markers are very efficient and informative in Opuntia species. Our results should be taken into consideration for plant breeding and genetic resource conservation programs. Further investigations are currently underway in order to determine the biochemical characteristics of cladode, flower, fruit and juice of these species, and to specify their molecular families to highlight the most appropriate use of each species.
Acknowledgements
We are thankful to Mr. Alfeddy Mohamed Najib (INRA, Marrakech) for providing us with the DNA thermal cycler.
References
- Abdel-Hameed E. S, Nagaty M. A, Salman M. S, Bazaid S. A. Phytochemicals, nutritionals and antioxidant properties of two prickly pear cactus cultivars (Opuntia ficus indica Mill.) growing in Taif, KSA. Food Chem; 16: 031–38: (2014)
- Alauddin M, Quiggin J. Agricultural intensification, irrigation and the environment in South Asia: Issues and policy options. Ecol Econ; 65: 111–124: (2008)
CrossRef - Andersen J. R, Lübberstedt T. Functional markers in plants. Trends Plant Sci; 8554-560: (2003)
CrossRef - Andrade-Montemayor H. M, Cordova-Torres A. V, García-Gasca T, Kawas J. R. Alternative foods for small ruminants in semiarid zones, the case of Mesquite (Prosopis laevigata spp.) and Nopal (Opuntia spp.). Small Rumin Res; 98: 83–92: (2011)
CrossRef - Arba M. Le cactus Opuntia, une espèce fruitière et fourragère pour une agriculture durable au Maroc. Symposium international agriculture durable en région méditerranéenne (AGDUMED), Rabat, Morocco; 215-223: (2009)
- Bendhifi M, Baraket G, Zourgui L, Souid S, Salhi-Hannachi A. Assessment of genetic diversity of Tunisian Barbary fig (Opuntia ficus indica) cultivars by RAPD markers and morphological traits. Sci Hortic; 158: 1-7: (2013)
CrossRef - Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Gen; 32: 314–331: (1980)
- Brake M, Migdadi H, Al-Gharaibeh M, Ayoub S, Haddad N, El Oqlah A. Characterization of Jordanian olive cultivars (Olea europaea L.) using RAPD and ISSR molecular markers. Sci Hortic; 176: 282-289: (2014)
CrossRef - Bretting P. K, Widrlechner M. P. Genetic markers and horticultural germplasm management. Hortic Sci; 30: 1349-1356: (1995)
- Caruso M, Curro S, Las Casas G, La Malfa S, Gentile A. Microsatellite markers help to assess genetic diversity among Opuntia ficus indica cultivated genotypes and their relation with related species. Plant Syst Evol; 290: 85–97: (2010)
CrossRef - Chapman B, Mondragón-Jacobo C, Bunch R. A, Paterson H. Breeding and Biotechnology. In: Nobel PS (ed) Cacti: Biology and Uses. University of California Press, California, USA; 255-271: (2002)
CrossRef - Charcosset A, Moreau L. Use of molecular markers for the development of new cultivars and the evaluation of genetic diversity. Euphytica; 137: 81–94: (2004)
CrossRef - Costa R. G, Treviño I. H, de Medeiros G. R, Medeiros A. N, Pinto T. F, de Oliveira R. L. Effects of replacing corn with cactus pear (Opuntia ficus indica Mill) on the performance of Santa Inês lambs. Small Rumin Res; 102: 13–17:(2012)
CrossRef - Demey J. R, Zambrano A. Y, Fuenmayor F, Segovia V. Relación entre caracterizaciones molecular y morfológica en una colección de yuca. Interciencia; 28: 684–689: (2003)
- Dereje M, Udén P. The browsing dromedary camel I. Behaviour, plant preference and quality of forage selected. Anim Feed Sci Technol; 121: 297–308: (2005)
CrossRef - Diederichsen A. Assessments of genetic diversity within a world collection of cultivated hexaploid oat (Avena sativa L.) based on qualitative morphological characters. Genet Resour Crop Evol; 55: 419-440: (2008)
CrossRef - Doyle J. J, Doyle J. L. Isolation of plant DNA from fresh tissue. Focus; 12: 13-15(1990)
- El Finti A, Belyadi M, El Boullani R, Msanda F, Serghini M. A, El Mousadik A. Genetic structure of cactus pear (Opuntia ficus indica) in Moroccan collection. Asian J Plant Sci; 12: 145-148: (2013)
CrossRef - Elhoumaizi M. A, Saaidi M, Oihabi A, Cilas C. Phenotypic diversity of date-palm cultivars (Phoenix dactylifera L.) from Morocco. Genet Resour Crop Evol; 49: 483-490: (2002)
CrossRef - El-Mostafa K, El Kharrassi Y, Badreddine A, Andreoletti P, Vamecq J, El Kebbaj M. S, Latruffe N, Lizard G, Nasser B, Cherkaoui-Malki M. Nopal cactus (Opuntia ficus-indica) as a source of bioactive compounds for nutrition, health and disease. Molecules;19: 14879-14901: (2014)
CrossRef - Erre P, Chessa I, Nieddu G, Jones P. G. Diversity and spatial distribution of Opuntia spp. in the Mediterranean Basin. J Arid Environ; 73: 1058-1066: (2009)
CrossRef - Felker P, del C Rodriguez S, Casoliba R. M, Filippini R, Medina D, Zapata R. Comparison of Opuntia ficus indica varieties of Mexican and Argentine origin for fruit yield and quality in Argentina. J Arid Environ; 60: 405-422: (2005)
CrossRef - Feugang J. M, Konarski P, Zou D, Stintzing F. C, Zou C. Nutritional and medicinal use of Cactus pear (Opuntia spp.) cladodes and fruits. Front Biosci; 11: 2574-2589: (2006)
CrossRef - Hartings H, Berardo N, Mazzinelli G. F, Valoti P, Verderio A, Motto M. Assessment of genetic diversity and relationships among maize (Zea mays L.) Italian landraces by morphological traits and AFLP profiling. Theor Appl Genet; 117: 831–842: (2008)
CrossRef - Helsen P, Browne R. A, Anderson D. J, Verdyck P, Van Dongen S. Galápagos’ Opuntia (prickly pear) cacti: extensive morphological diversity, low genetic variability. Biol J Linnean Soc; 96: 451–461: (2009)
CrossRef - Kölliker R, Jones E. S, Jahufer M. Z. Z, Forster J. W. Bulked AFLP analysis for the assessment of genetic diversity in white clover (Trifolium repens L.). Euphytica; 121: 305-315: (2001)
CrossRef - Labra M, Grassi F, Bardini M, Imazio S, Guiggi A, Citterio S, Banfi E, Sgorbati S. Genetic relationships in Opuntia Mill. genus (Cactaceae) detected by molecular marker. Plant Sci; 165: 1129-1136: (2003)
CrossRef - Lee M. DNA markers and plant breeding programs. Adv Agron; 55: 265–344: (1995)
CrossRef - Malik S. K, Rohini M. R, Kumar S, Choudhary R, Pal D, Chaudhury R. Assessment of genetic diversity in sweet orange [Citrus sinensis (L.) Osbeck] cultivars of India using morphological and RAPD markers. Agri Res; 1: 317–324: (2012)
CrossRef - Mohamed-Yasseen Y, Barringer S. A, Splittstoesser W. E. A note on the uses of Opuntia spp. in Central/North America. J Arid Environ; 32: 347 –353: (1996)
CrossRef - Peña-Valdivia C. B, Luna-Cavazos M, Carranza-Sabas J. A, Reyes-Agüero J. A, Flores A. Morphological characterization of Opuntia spp.: a multivariate analysis. J Prof Assoc Cactus Dev; 10: 1-21. (2008)
- Piga A. Cactus pear: a fruit of nutraceutical and functional importance. J Prof Assoc Cactus Dev; 6: 9-22: (2004)
- Realini M. F, Gonzalez G. E, Font F, Picca P. I, Poggio L, Gottlieb A. M. Phylogenetic relationships in Opuntia (Cactaceae, Opuntioideae) from southern South America. Plant Syst Evol; doi: 10.1007/s00606-014-1154-1: (2014)
CrossRef - Rohlf F. J. NTSYSpc numerical taxonomy and multivariate analysis system version 2.1, users guide. Exeter Software. Setauket, New York; (2000)
- Sedra M. H, Lashermes P, Trouslot P, Combes M.C, Hamon S. Identification and genetic diversity analysis of date palm (Phoenix dactylifera L.) varieties from Morocco using RAPD markers. Euphytica; 103: 75–82: (1998)
CrossRef - Semagn K, Bjørnstad A, Ndjiondjop M. N. An overview of molecular marker methods for plants. Afr J Biotechnol; 5: 2540-2568: (2006)
- Shahnejat-Bushehri A. A, Torabi S, Omidi M, Ghannadha M. R. Comparison of Genetic and Morphological Distance with Heterosis with RAPD Markers in Hybrids of Barley. Int J Agri Biol; 7592-595: (2005)
- Talebi R, Fayyaz F. Quantitative evaluation of genetic diversity in Iranian modern cultivars of wheat (Triticum aestivum L.) using morphological and amplified fragment length polymorphism (AFLP) markers. Biharean Biol; 6: 14-18: (2012)
- Valadez-Moctezuma E, Samah S, Luna-Paez A. Genetic diversity of Opuntia spp. varieties assessed by classical marker tools (RAPD and ISSR). Plant Syst Evol; doi: 10.1007/s00606-014-1112-y: (2014)
CrossRef - West-Eberhard M. J. Phenotypic plasticity and the origins of diversity. Ann Rev Ecol Syst; 20: 249-278: (1989)
CrossRef - Williams J. G. K, Kubelik A. R, Livak K. J, Rafalski J. A, Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res; 18: 6531 – 6535: (1990)
CrossRef - Yahia E. M, Mondragon-Jacobo C. Nutritional components and anti-oxidant capacity of ten cultivars and lines of cactus pear fruit (Opuntia spp.). Food Res Int; 44: 2311–2318: (2011)
CrossRef - Yawen, Z, Shiquan S, Zichao L, Zhongyi Y, Xiangkun W, Hongliang Z, Guosong W. Ecogeographic and genetic diversity based on morphological characters of indigenous rice (Oryza sativa L.) in Yunnan, China. Genet Resour Crop Evol; 50: 567–577: (2003)
CrossRef - Zoghlami N, Chrita I, Bouamama B, Gargouri M, Zemni H, Ghorbel A, Mliki A. Molecular based assessment of genetic diversity within Barbary fig (Opuntia ficus indica (L.) Mill.) in Tunisia. Sci Hortic; 113: 134-141: (2007)
CrossRef
